Workshop Description
The recount2 project re-processed RNA sequencing (RNA-seq) data on over 70,000 human RNA-seq samples spanning a variety of tissues, cell types and disease conditions. Researchers can easily access these data via the recount Bioconductor package, and can quickly import gene, exon, exon-exon junction and base-pair coverage data for uniformly processed data from the SRA, GTEx and TCGA projects in R for analysis. This workshop will include an overview of the recount2 project as well as methods and tools that have been to improve it since 2017. The overview will be followed by a hands-on session where you will dive into recount and related packages along the lines of the recount workflow. We will also cover new related packages such as GenomicState.
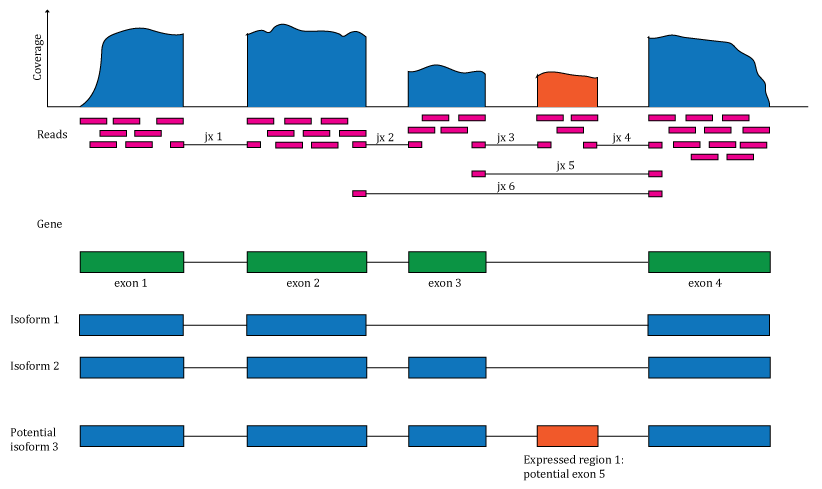
In more detail, this workshop will cover different use cases of the recount package, including downloading and normalizing data, processing and cleaning relevant phenotype data (including recount-brain), performing differential expression (DE) analyses using other Bioconductor packages. The workshop will also cover how to use the base-pair coverage data for annotation-agnostic DE analyses and for visualizing coverage data for features of interest. After taking this workshop, attendees will be ready to enhance their analyses by leveraging RNA-seq data from 70,000 human samples.
Pre-requisites
- Basic knowledge of R syntax
- Basic knowledge of RNA-seq or gene expression data
- Familiarity with
SummarizedExperimentvignette - Familiarity with the
RangedSummarizedExperimentclass
Workshop Participation
Students are expected to ask questions during the recount2 overview presentation and to bring their own laptop so they can follow the hands-on portion of the workshop.
The slides for the workshop are available through speakerdeck.
You can download a Docker image with all the workshop files using:
Then, log in to RStudio at http://localhost:8787 using username rstudio and password bioc2020. Note that on Windows you need to provide your localhost IP address like http://191.163.92.108:8787/ - find it using docker-machine ip default in Docker’s terminal.
To see the vignette on RStudio’s window (from the docker image), run browseVignettes(package = "recountWorkshop2020"). Click on one of the links, “HTML”, “source”, “R code”. In case of The requested page was not found error, add help/ to the URL right after the hostname, e.g., http://localhost:8787/help/library/recountWorkshop2020/doc/recount-workshop.html. This is a known bug.
Workshop goals and objectives
Installation instructions
Get the latest stable R release from CRAN. Then install recountWorkshop2020 using from GitHub with the following code:
if (!requireNamespace("BiocManager", quietly = TRUE)) { install.packages("BiocManager") } BiocManager::install("LieberInstitute/recountWorkshop2020")
Citation
Below is the citation output from using citation('recount in R. Please run this yourself to check for any updates on how to cite recount.
print(citation("recount")[2], bibtex = TRUE) #> #> Collado-Torres L, Nellore A, Jaffe AE (2017). "recount workflow: #> Accessing over 70,000 human RNA-seq samples with Bioconductor [version #> 1; referees: 1 approved, 2 approved with reservations]." #> _F1000Research_. doi: 10.12688/f1000research.12223.1 (URL: #> https://doi.org/10.12688/f1000research.12223.1), <URL: #> https://f1000research.com/articles/6-1558/v1>. #> #> A BibTeX entry for LaTeX users is #> #> @Article{, #> title = {recount workflow: Accessing over 70,000 human RNA-seq samples with Bioconductor [version 1; referees: 1 approved, 2 approved with reservations]}, #> author = {Leonardo Collado-Torres and Abhinav Nellore and Andrew E. Jaffe}, #> year = {2017}, #> journal = {F1000Research}, #> doi = {10.12688/f1000research.12223.1}, #> url = {https://f1000research.com/articles/6-1558/v1}, #> }
Please note that the recount was only made possible thanks to many other R and bioinformatics software authors, which are cited either in the vignettes and/or the paper(s) describing this package.
Code of Conduct
Please note that the recountWorkshop2020 project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Development tools
- Continuous code testing is possible thanks to GitHub actions through usethis, remotes, sysreqs and rcmdcheck customized to use Bioconductor’s docker containers and BiocCheck.
- Code coverage assessment is possible thanks to codecov and covr.
- The documentation website is automatically updated thanks to pkgdown.
- The code is styled automatically thanks to styler.
- The documentation is formatted thanks to devtools and roxygen2.
For more details, check the dev directory.
This package was developed using biocthis.
Links
- Report a bug at
https://support.bioconductor.org/t/recountWorkshop2020
License
Community
Citation
Developers
- Leonardo Collado-Torres
Author, maintainer - All authors...
