4 Pipeline Overview
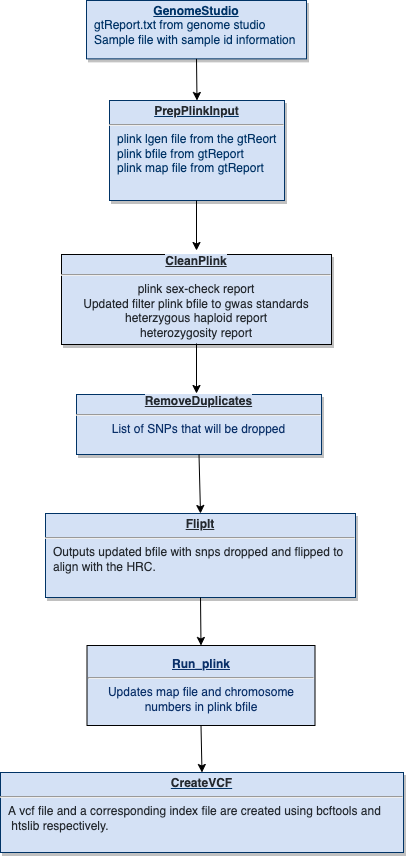
Diagram representing the “conceptual” workflow traversed by PopTop. Here some nextflow processes are grouped together for simplicity; the exact processes traversed are enumerated below.
4.1 Main Workflow Steps
- PrepPlinkInput: reformats a GenomeStudio gtReport.txt in corresponding plink files.
- CleanPlink: The plink files are filtered by gwas standards and made into plink .bed files.
- RemoveDuplicates: duplicated snps that we found in the previous process are dropped from the genotypes.
- FlipIt: the first step of this process is to flip the snps to all be on the same strand. Multi-allelic snps will be dropped here. In the last step of this process the genotype data is aligned to the hrc to be incompliance with the TopMed imputation server standards.
- RunPlink: Drops snps that were not resolved in the HRC alignment. Chromosome format is updated and along with map with file being updated. Any remaining mismatches are flipped. Reference alleles are flipped to align with the hrc.
- CreateVCF: takes plink files and converts them to vcf format with a corresponding index file using bcftools.