Create low-res images and scale factors from high-res Fiji output images
Source:R/prep_fiji_image.R
prep_fiji_image.RdAfter stitching all groups in sample_info with Fiji, images of
various resolutions (pixel dimensions) are left. This function creates copies
of each image whose largest dimension is lowres_max_size pixels. It
also creates a corresponding scalefactors_json.json file much like
SpaceRanger's. In conjunction with prep_fiji_coords(), this function
prepares for building the SpatialExperiment-class
with build_spe().
prep_fiji_image(sample_info, out_dir, lowres_max_size = 1200)Arguments
- sample_info
A
data.frame()with columnscapture_area,group,fiji_xml_path,fiji_image_path,spaceranger_dir,intra_group_scalar, andgroup_hires_scalef. The last two are made byrescale_fiji_inputs().- out_dir
A
character(1)vector giving a path to a directory to place the output image(s) and scale factors. Provided the parent directory exists,out_dirwill be created if necessary.- lowres_max_size
An
integer(1)vector: the resolution (number of pixels) of the larger dimension of the output image(s), considered to be "low resolution". The default value of1200assumes that you are stitching together at most a 2 by 2 grid of Visium capture areas, where each has at most 600 pixels on the longest dimension (as is the default in SpaceRanger).
Value
This function returns character() with the file paths to the
tissue_lowres_image.png and scalefactors_json.json files it created.
Examples
if (file.exists("sample_info.rds")) {
sample_info <- readRDS("sample_info.rds")
} else {
sample_info <- dplyr::tibble(
group = "Br2719",
capture_area = c("V13B23-283_A1", "V13B23-283_C1", "V13B23-283_D1")
)
# Add 'spaceranger_dir' column
sr_dir <- tempdir()
temp <- unzip(
spatialLIBD::fetch_data("visiumStitched_brain_spaceranger"),
exdir = sr_dir
)
sample_info$spaceranger_dir <- file.path(
sr_dir, sample_info$capture_area, "outs", "spatial"
)
# Add Fiji-output-related columns
fiji_dir <- tempdir()
temp <- unzip(
spatialLIBD::fetch_data("visiumStitched_brain_Fiji_out"),
exdir = fiji_dir
)
sample_info$fiji_xml_path <- temp[grep("xml$", temp)]
sample_info$fiji_image_path <- temp[grep("png$", temp)]
## Re-size images and add more information to the sample_info
sample_info <- rescale_fiji_inputs(sample_info, out_dir = tempdir())
saveRDS(sample_info, "sample_info.rds")
}
spe_input_dir <- tempdir()
out_paths <- prep_fiji_image(
sample_info,
out_dir = spe_input_dir, lowres_max_size = 1000
)
# A "low resolution" stitched image was produced, which has 1000
# pixels in its largest dimension
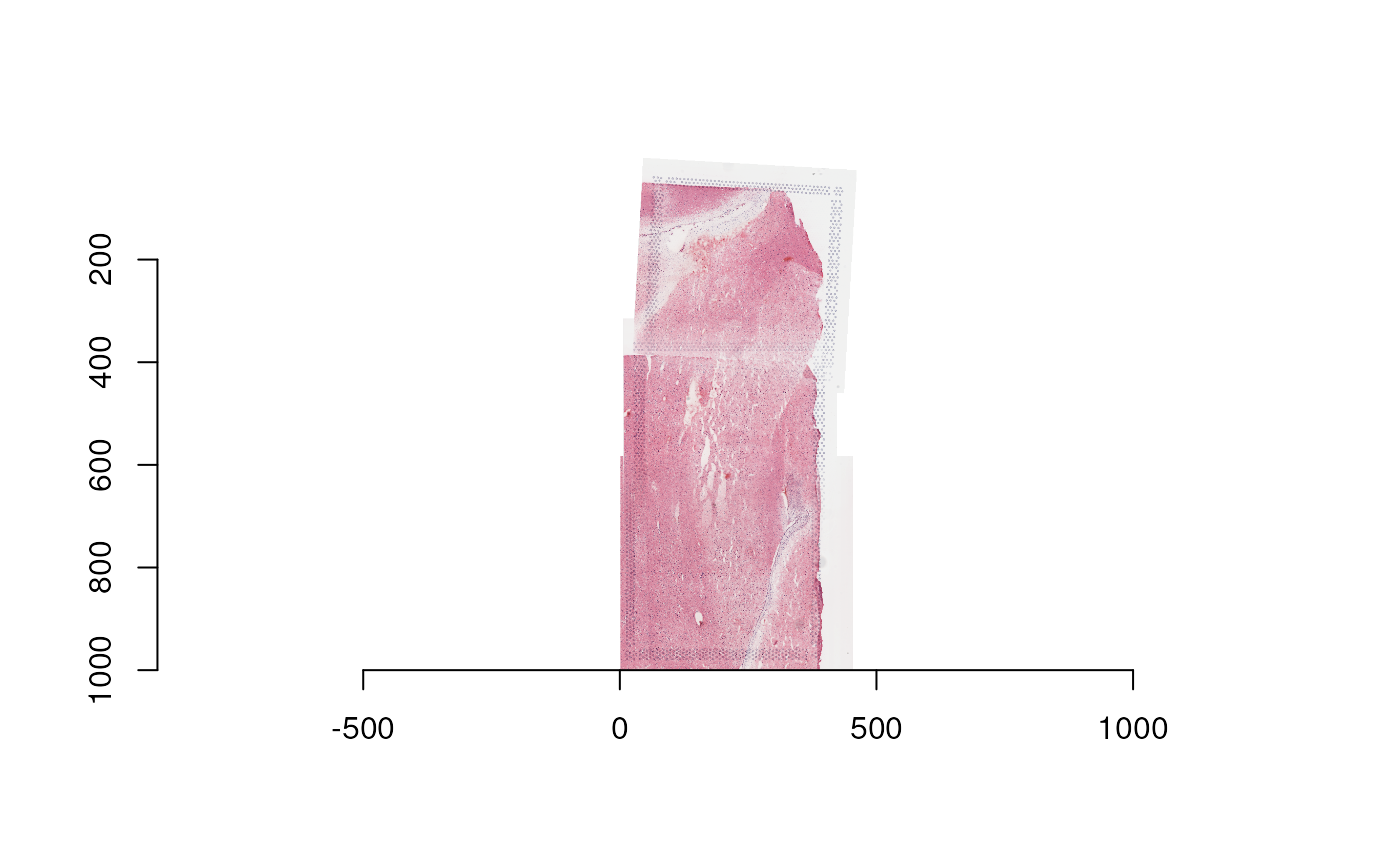
this_image <- imager::load.image(
file.path(spe_input_dir, "Br2719", "tissue_lowres_image.png")
)
dim(this_image)
#> [1] 461 1000 1 3
library("imager")
#> Loading required package: magrittr
#>
#> Attaching package: ‘magrittr’
#> The following object is masked from ‘package:GenomicRanges’:
#>
#> subtract
#>
#> Attaching package: ‘imager’
#> The following object is masked from ‘package:magrittr’:
#>
#> add
#> The following objects are masked from ‘package:SummarizedExperiment’:
#>
#> resize, width
#> The following object is masked from ‘package:Biobase’:
#>
#> channel
#> The following objects are masked from ‘package:GenomicRanges’:
#>
#> resize, width
#> The following objects are masked from ‘package:IRanges’:
#>
#> resize, width
#> The following object is masked from ‘package:S4Vectors’:
#>
#> width
#> The following object is masked from ‘package:BiocGenerics’:
#>
#> width
#> The following objects are masked from ‘package:stats’:
#>
#> convolve, spectrum
#> The following object is masked from ‘package:graphics’:
#>
#> frame
#> The following object is masked from ‘package:base’:
#>
#> save.image
plot(this_image)
 # In total, an image and scalefactors were written
out_paths
#> [1] "/tmp/RtmpAA9v23/Br2719/tissue_lowres_image.png"
#> [2] "/tmp/RtmpAA9v23/Br2719/scalefactors_json.json"
# In total, an image and scalefactors were written
out_paths
#> [1] "/tmp/RtmpAA9v23/Br2719/tissue_lowres_image.png"
#> [2] "/tmp/RtmpAA9v23/Br2719/scalefactors_json.json"