BiocMAP
Lieber Institute for Brain Development, Johns Hopkins Medical CampusLieber Institute for Brain Development, Johns Hopkins Medical CampusCenter for Computational Biology, Johns Hopkins University
lcolladotor@gmail.com
Overview
BiocMAP is a pair of Nextflow-based pipelines for processing raw whole genome bisulfite sequencing data into Bioconductor-friendly bsseq objects in R.
The first BiocMAP module performs speedy alignment to a reference genome by Arioc, and requires GPU resources. Methylation extraction and remaining steps are performed in the second module, optionally on a different computing system where GPUs need not be available.
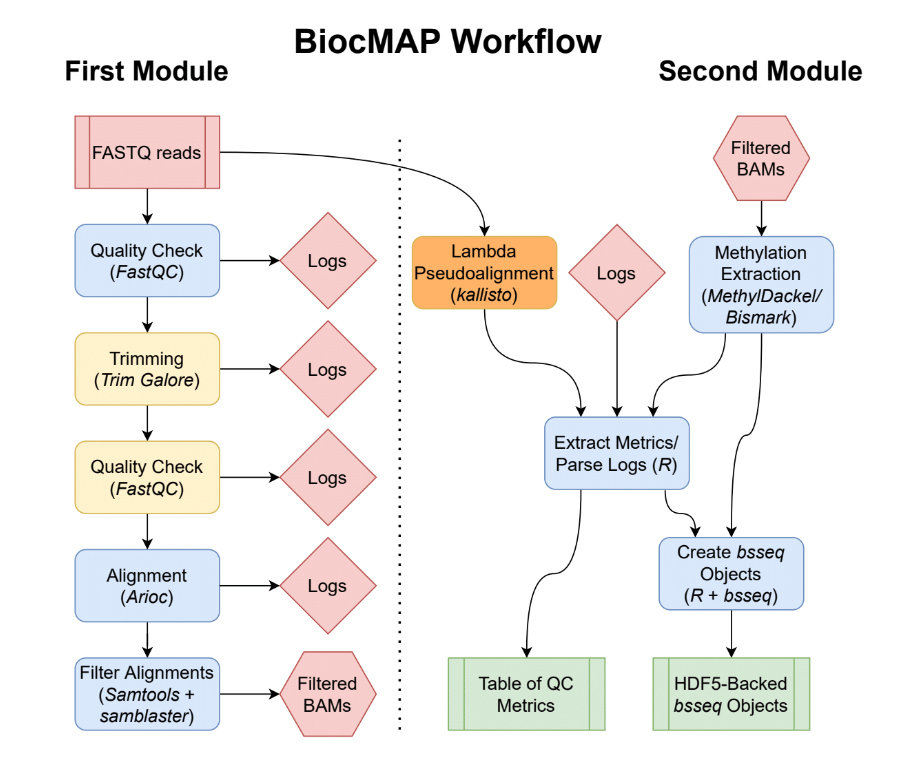 Diagram representing the “conceptual” workflow traversed by BiocMAP. Here some nextflow
Diagram representing the “conceptual” workflow traversed by BiocMAP. Here some nextflow processes are grouped together for simplicity; the exact processes traversed are enumerated here. The red box indicates the FASTQ files are inputs to the pipeline; green coloring denotes major output files from the pipeline; the remaining boxes represent computational steps. Yellow-colored steps are optional or not always performed; for example, lambda pseudoalignment is an optional step intended for experiments with spike-ins of the lambda bacteriophage. Finally, blue-colored steps are ordinary processes which occur on every pipeline execution.
Cite BiocMAP
We hope BiocMAP will be a useful tool for your research. Please use the following bibtex information to cite this software. Thank you!
@article {Eagles2022.04.20.488947,
author = {Eagles, Nicholas J and Wilton, Richard and Jaffe, Andrew E. and Collado-Torres, Leonardo},
title = {BiocMAP: A Bioconductor-friendly, GPU-Accelerated Pipeline for Bisulfite-Sequencing Data},
year = {2022},
doi = {10.1101/2022.04.20.488947},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://doi.org/10.1101/2022.04.20.488947},
journal = {bioRxiv}
}This is a project by the R/Bioconductor-powered Team Data Science at the Lieber Institute for Brain Development.
 |
 |
R session information
Details on the R version used for making this book. The source code is available at LieberInstitute/BiocMAP.
## Load the package at the top of your script
library("sessioninfo")
## Reproducibility information
print('Reproducibility information:')
Sys.time()
proc.time()
options(width = 120)
session_info()## [1] "Reproducibility information:"## [1] "2025-12-10 20:01:57 UTC"## user system elapsed
## 0.458 0.348 0.444## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R Under development (unstable) (2025-12-07 r89119)
## os Ubuntu 24.04.3 LTS
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz UTC
## date 2025-12-10
## pandoc 3.8.3 @ /usr/bin/ (via rmarkdown)
## quarto 1.7.32 @ /usr/local/bin/quarto
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## bookdown 0.46 2025-12-05 [1] CRAN (R 4.6.0)
## bslib 0.9.0 2025-01-30 [2] CRAN (R 4.6.0)
## cachem 1.1.0 2024-05-16 [2] CRAN (R 4.6.0)
## cli 3.6.5 2025-04-23 [2] CRAN (R 4.6.0)
## digest 0.6.39 2025-11-19 [2] CRAN (R 4.6.0)
## evaluate 1.0.5 2025-08-27 [2] CRAN (R 4.6.0)
## fastmap 1.2.0 2024-05-15 [2] CRAN (R 4.6.0)
## htmltools 0.5.9 2025-12-04 [2] CRAN (R 4.6.0)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.6.0)
## jsonlite 2.0.0 2025-03-27 [2] CRAN (R 4.6.0)
## knitr 1.50 2025-03-16 [2] CRAN (R 4.6.0)
## lifecycle 1.0.4 2023-11-07 [2] CRAN (R 4.6.0)
## R6 2.6.1 2025-02-15 [2] CRAN (R 4.6.0)
## rlang 1.1.6 2025-04-11 [2] CRAN (R 4.6.0)
## rmarkdown 2.30 2025-09-28 [2] CRAN (R 4.6.0)
## rstudioapi 0.17.1 2024-10-22 [2] CRAN (R 4.6.0)
## sass 0.4.10 2025-04-11 [2] CRAN (R 4.6.0)
## sessioninfo * 1.2.3 2025-02-05 [2] CRAN (R 4.6.0)
## xfun 0.54 2025-10-30 [2] CRAN (R 4.6.0)
## yaml 2.3.11 2025-11-28 [2] CRAN (R 4.6.0)
##
## [1] /usr/local/lib/R/host-site-library
## [2] /usr/local/lib/R/site-library
## [3] /usr/local/lib/R/library
## * ── Packages attached to the search path.
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────This book was last updated on 2025-12-10 20:01:57.284813.