Hacking our way through UpSetR
For our club meeting today we were going to summarize the Demystifying Data Science conference but we forgot that the videos are not released yet.
Oops, we'll have to postpone our blog post. We didn't read the fine print that talk recordings will be available sometime next week. Sorry about that!
— LIBD rstats club (@LIBDrstats) July 27, 2018
So we adjusted plans and decided to continue our work on the UpSetR (Gehlenborg, 2016) package by Nils Gehlenborg.
Yesterday we discussed various options for visualizing large numbers of overlapping groups. We explored uses of the #venneuler package from Lee Wilkinson and Simon Urbanek and the #UpSetR package from Jake Conway, Alexander Lex, and @ngehlenborg. #rstats pic.twitter.com/k55YfihmiP
— LIBD rstats club (@LIBDrstats) June 16, 2018
What you can currently do
First, let’s install the version we used for this post:
devtools::install_github('hms-dbmi/UpSetR@fe2812c')Our ultimate goal is to submit a pull request that enables UpSetR users to specify a color by row for the dots instead of the actual rows. We had already identified an example that we could work with.
library('UpSetR')
movies <- read.csv( system.file("extdata", "movies.csv", package = "UpSetR"),
header=T, sep=";" )
require(ggplot2); require(plyr); require(gridExtra); require(grid);## Loading required package: ggplot2## Loading required package: plyr## Loading required package: gridExtra## Loading required package: gridupset(movies,
sets = c("Action", "Comedy", "Drama"),
order.by="degree", matrix.color="blue", point.size=5,
sets.bar.color=c("maroon","blue","orange"))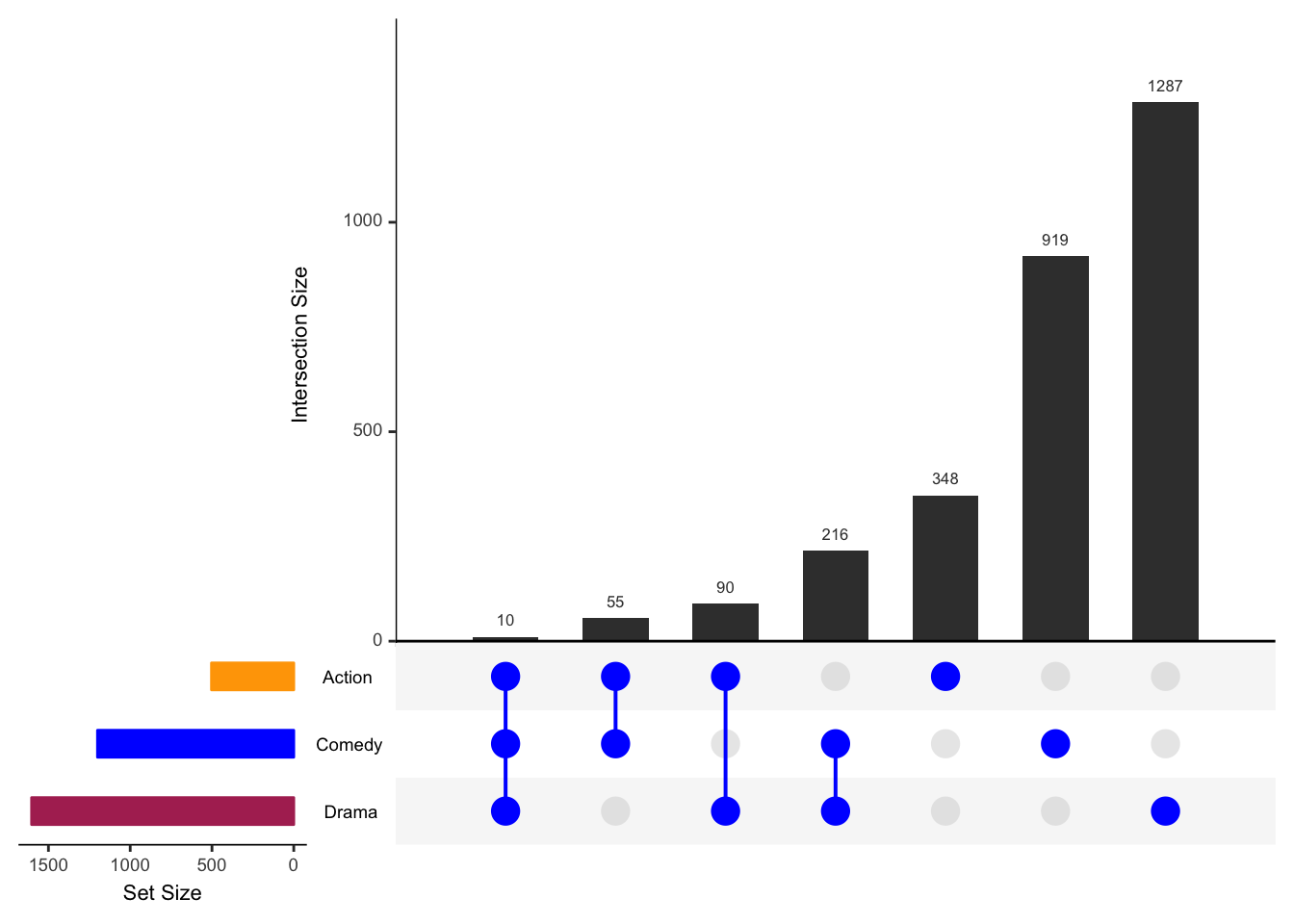
We also explored the set metadata vignette that includes examples such as the following one.
set.seed(20180727)
## Create the metadata object first
sets <- names(movies[3:19])
avgRottenTomatoesScore <- round(runif(17, min = 0, max = 90))
metadata <- as.data.frame(cbind(sets, avgRottenTomatoesScore))
names(metadata) <- c("sets", "avgRottenTomatoesScore")
metadata$avgRottenTomatoesScore <- as.numeric(as.character(metadata$avgRottenTomatoesScore))
Cities <- sample(c("Boston", "NYC", "LA"), 17, replace = T)
metadata <- cbind(metadata, Cities)
metadata$Cities <- as.character(metadata$Cities)
metadata[which(metadata$sets %in% c("Drama", "Comedy", "Action", "Thriller",
"Romance")), ]## sets avgRottenTomatoesScore Cities
## 1 Action 68 Boston
## 4 Comedy 40 NYC
## 7 Drama 48 LA
## 13 Romance 77 Boston
## 15 Thriller 19 NYCaccepted <- round(runif(17, min = 0, max = 1))
metadata <- cbind(metadata, accepted)
metadata[which(metadata$sets %in% c("Drama", "Comedy", "Action", "Thriller",
"Romance")), ]## sets avgRottenTomatoesScore Cities accepted
## 1 Action 68 Boston 0
## 4 Comedy 40 NYC 1
## 7 Drama 48 LA 0
## 13 Romance 77 Boston 1
## 15 Thriller 19 NYC 0## Now make the plot
upset(movies, set.metadata = list(data = metadata, plots = list(list(type = "hist",
column = "avgRottenTomatoesScore", assign = 20), list(type = "matrix_rows",
column = "Cities", colors = c(Boston = "green", NYC = "navy", LA = "purple"),
alpha = 0.5))))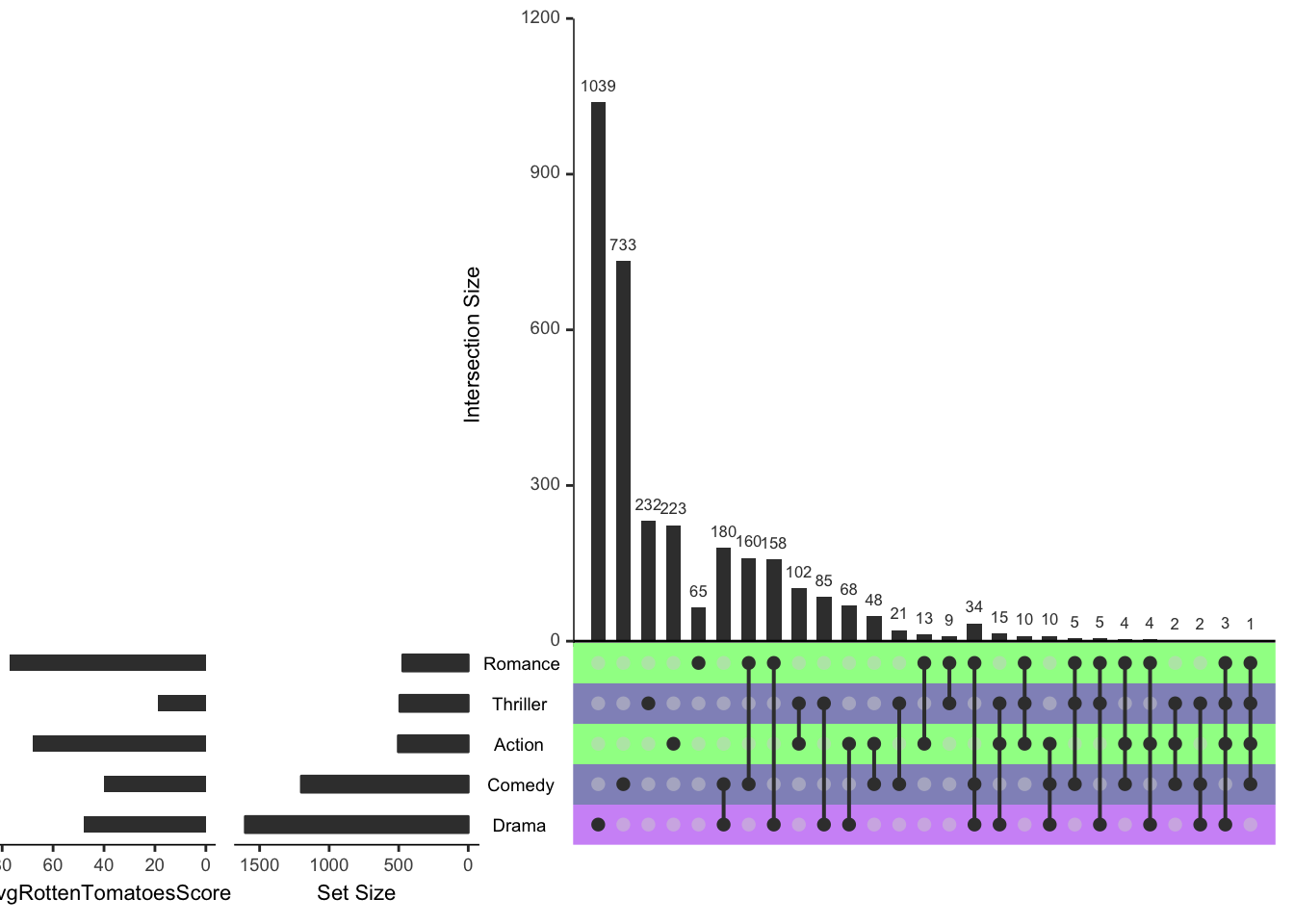
Hacking our way
Using the metadata looked complicated to us and hopefully not necessary for what we are trying to accomplish. That is, we really wanted to change the colors of the circles in each row, not the rows themselves. So we found the GitHub repo with the code, plugged a laptop to a TV and started exploring as a group. We went the rabbit hole to see how the matrix.color argument got used. To actually hack our way through, we downloaded the latest version of the code using git.
git clone git@github.com:hms-dbmi/UpSetR.git
cd UpSetRNext, we created the objects that match the default arguments of upset() by finding and replacing commas by semi-colons. Well, not all of the commas. Also, for inputs that specified a vector (mostly 2 options), we chose the first one to match the default R behavior. This way we could execute them and have them in our session.
## Default upset() arguments
nsets = 5; nintersects = 40; sets = NULL; keep.order = F; set.metadata = NULL; intersections = NULL;
matrix.color = "gray23"; main.bar.color = "gray23"; mainbar.y.label = "Intersection Size"; mainbar.y.max = NULL;
sets.bar.color = "gray23"; sets.x.label = "Set Size"; point.size = 2.2; line.size = 0.7;
mb.ratio = c(0.70,0.30); expression = NULL; att.pos = NULL; att.color = main.bar.color; order.by = 'freq';
decreasing = T; show.numbers = "yes"; number.angles = 0; group.by = "degree";cutoff = NULL;
queries = NULL; query.legend = "none"; shade.color = "gray88"; shade.alpha = 0.25; matrix.dot.alpha =0.5;
empty.intersections = NULL; color.pal = 1; boxplot.summary = NULL; attribute.plots = NULL; scale.intersections = "identity";
scale.sets = "identity"; text.scale = 1; set_size.angles = 0 ; set_size.show = FALSE Next, we did the same (commas to semicolons) for the inputs of the first example.
## Initial inputs on the first example
movies <- read.csv( system.file("extdata", "movies.csv", package = "UpSetR"),
header=T, sep=";" )
## comma -> semicolon
data = movies; sets = c("Action", "Comedy", "Drama");
order.by="degree"; matrix.color="blue"; point.size=5;
sets.bar.color=c("maroon","blue","orange")Now we were ready to start modifying some of the internal UpSetR (Gehlenborg, 2016) code.
Hacking internals
The function upset() is pretty long and uses many un-exported functions from the package itself. In order to test thing quickly we added UpSetR::: calls before the un-exported functions. Here’s our modified version where we added a piece of code to modify the Matrix_layout object and add some colors.
## Piece of code we introduced
for(i in 1:3) {
j <- which(Matrix_layout$y == i & Matrix_layout$value == 1)
if(length(j) > 0) Matrix_layout$color[j] <- c("maroon","blue","orange")[i]
}Ok, here’s the full modified upset() function.
## Modified internal upset() code
startend <- UpSetR:::FindStartEnd(data)
first.col <- startend[1]
last.col <- startend[2]
if(color.pal == 1){
palette <- c("#1F77B4", "#FF7F0E", "#2CA02C", "#D62728", "#9467BD", "#8C564B", "#E377C2",
"#7F7F7F", "#BCBD22", "#17BECF")
} else{
palette <- c("#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00",
"#CC79A7")
}
if(is.null(intersections) == F){
Set_names <- unique((unlist(intersections)))
Sets_to_remove <- UpSetR:::Remove(data, first.col, last.col, Set_names)
New_data <- UpSetR:::Wanted(data, Sets_to_remove)
Num_of_set <- UpSetR:::Number_of_sets(Set_names)
if(keep.order == F){
Set_names <- UpSetR:::order_sets(New_data, Set_names)
}
All_Freqs <- UpSetR:::specific_intersections(data, first.col, last.col, intersections, order.by, group.by, decreasing,
cutoff, main.bar.color, Set_names)
} else if(is.null(intersections) == T){
Set_names <- sets
if(is.null(Set_names) == T || length(Set_names) == 0 ){
Set_names <- UpSetR:::FindMostFreq(data, first.col, last.col, nsets)
}
Sets_to_remove <- UpSetR:::Remove(data, first.col, last.col, Set_names)
New_data <- UpSetR:::Wanted(data, Sets_to_remove)
Num_of_set <- UpSetR:::Number_of_sets(Set_names)
if(keep.order == F){
Set_names <- UpSetR:::order_sets(New_data, Set_names)
}
All_Freqs <- UpSetR:::Counter(New_data, Num_of_set, first.col, Set_names, nintersects, main.bar.color,
order.by, group.by, cutoff, empty.intersections, decreasing)
}
Matrix_setup <- UpSetR:::Create_matrix(All_Freqs)
labels <- UpSetR:::Make_labels(Matrix_setup)
#Chose NA to represent NULL case as result of NA being inserted when at least one contained both x and y
#i.e. if one custom plot had both x and y, and others had only x, the y's for the other plots were NA
#if I decided to make the NULL case (all x and no y, or vice versa), there would have been alot more if/else statements
#NA can be indexed so that we still get the non NA y aesthetics on correct plot. NULL cant be indexed.
att.x <- c(); att.y <- c();
if(is.null(attribute.plots) == F){
for(i in seq_along(attribute.plots$plots)){
if(length(attribute.plots$plots[[i]]$x) != 0){
att.x[i] <- attribute.plots$plots[[i]]$x
}
else if(length(attribute.plots$plots[[i]]$x) == 0){
att.x[i] <- NA
}
if(length(attribute.plots$plots[[i]]$y) != 0){
att.y[i] <- attribute.plots$plots[[i]]$y
}
else if(length(attribute.plots$plots[[i]]$y) == 0){
att.y[i] <- NA
}
}
}
BoxPlots <- NULL
if(is.null(boxplot.summary) == F){
BoxData <- UpSetR:::IntersectionBoxPlot(All_Freqs, New_data, first.col, Set_names)
BoxPlots <- list()
for(i in seq_along(boxplot.summary)){
BoxPlots[[i]] <- UpSetR:::BoxPlotsPlot(BoxData, boxplot.summary[i], att.color)
}
}
customAttDat <- NULL
customQBar <- NULL
Intersection <- NULL
Element <- NULL
legend <- NULL
EBar_data <- NULL
if(is.null(queries) == F){
custom.queries <- UpSetR:::SeperateQueries(queries, 2, palette)
customDat <- UpSetR:::customQueries(New_data, custom.queries, Set_names)
legend <- UpSetR:::GuideGenerator(queries, palette)
legend <- UpSetR:::Make_legend(legend)
if(is.null(att.x) == F && is.null(customDat) == F){
customAttDat <- UpSetR:::CustomAttData(customDat, Set_names)
}
customQBar <- UpSetR:::customQueriesBar(customDat, Set_names, All_Freqs, custom.queries)
}
if(is.null(queries) == F){
Intersection <- UpSetR:::SeperateQueries(queries, 1, palette)
Matrix_col <- UpSetR:::intersects(QuerieInterData, Intersection, New_data, first.col, Num_of_set,
All_Freqs, expression, Set_names, palette)
Element <- UpSetR:::SeperateQueries(queries, 1, palette)
EBar_data <-UpSetR:::ElemBarDat(Element, New_data, first.col, expression, Set_names,palette, All_Freqs)
} else{
Matrix_col <- NULL
}
Matrix_layout <- UpSetR:::Create_layout(Matrix_setup, matrix.color, Matrix_col, matrix.dot.alpha)As a little pause in upset(), let’s check what actually Matrix_layout looks.
Matrix_layout## y x value color alpha Intersection
## 1 1 1 1 blue 1.0 1yes
## 2 2 1 1 blue 1.0 1yes
## 3 3 1 1 blue 1.0 1yes
## 4 1 2 0 gray83 0.5 4No
## 5 2 2 1 blue 1.0 2yes
## 6 3 2 1 blue 1.0 2yes
## 7 1 3 1 blue 1.0 3yes
## 8 2 3 0 gray83 0.5 8No
## 9 3 3 1 blue 1.0 3yes
## 10 1 4 1 blue 1.0 4yes
## 11 2 4 1 blue 1.0 4yes
## 12 3 4 0 gray83 0.5 12No
## 13 1 5 0 gray83 0.5 13No
## 14 2 5 0 gray83 0.5 14No
## 15 3 5 1 blue 1.0 5yes
## 16 1 6 0 gray83 0.5 16No
## 17 2 6 1 blue 1.0 6yes
## 18 3 6 0 gray83 0.5 18No
## 19 1 7 1 blue 1.0 7yes
## 20 2 7 0 gray83 0.5 20No
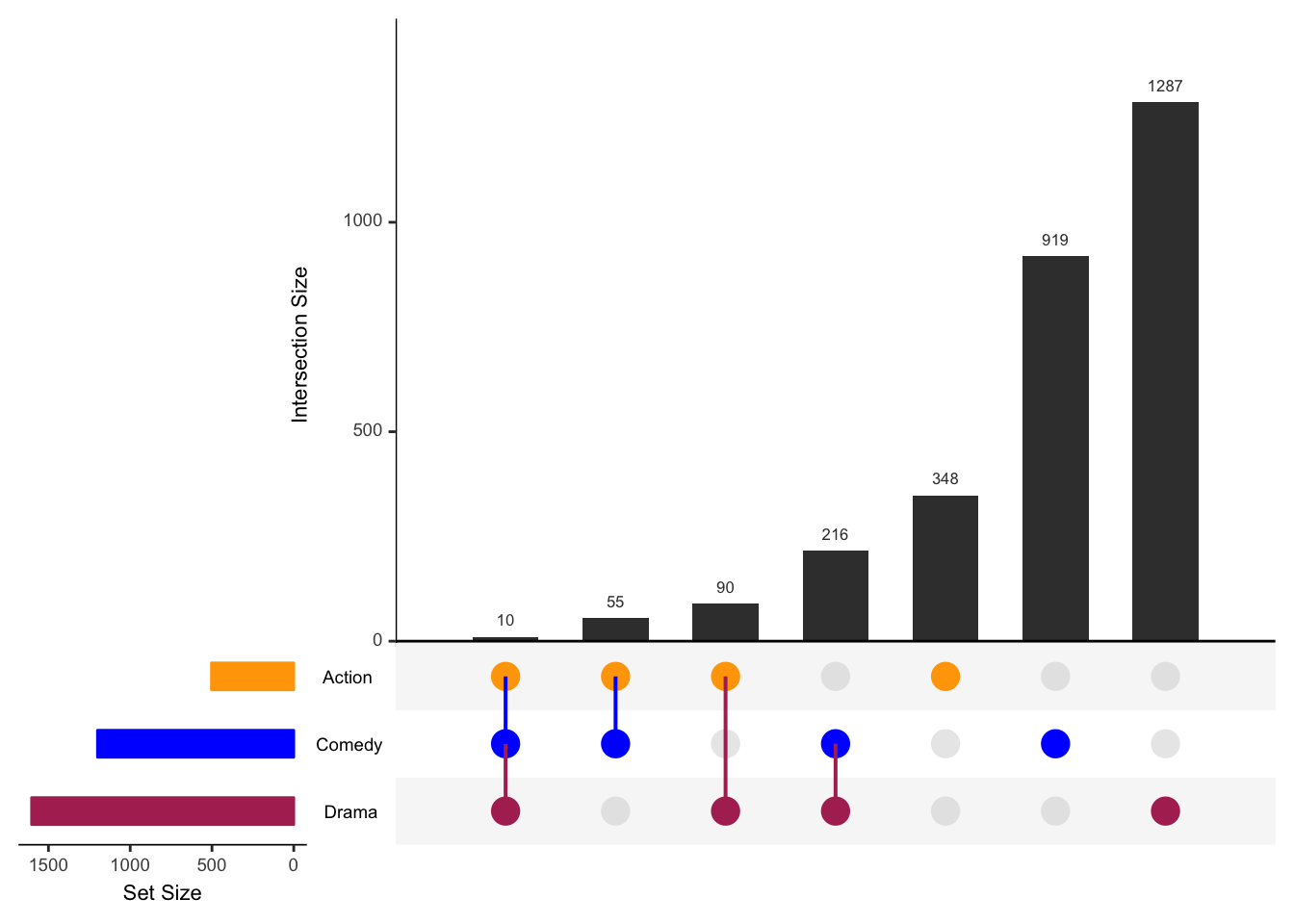
## 21 3 7 0 gray83 0.5 21NoWe figured out that we had to change the colors only the rows with value = 1 and that y was the row grouping variable.
## our modification
for(i in 1:3) {
j <- which(Matrix_layout$y == i & Matrix_layout$value == 1)
if(length(j) > 0) Matrix_layout$color[j] <- c("maroon","blue","orange")[i]
}Here’s our modified Matrix_layout:
Matrix_layout## y x value color alpha Intersection
## 1 1 1 1 maroon 1.0 1yes
## 2 2 1 1 blue 1.0 1yes
## 3 3 1 1 orange 1.0 1yes
## 4 1 2 0 gray83 0.5 4No
## 5 2 2 1 blue 1.0 2yes
## 6 3 2 1 orange 1.0 2yes
## 7 1 3 1 maroon 1.0 3yes
## 8 2 3 0 gray83 0.5 8No
## 9 3 3 1 orange 1.0 3yes
## 10 1 4 1 maroon 1.0 4yes
## 11 2 4 1 blue 1.0 4yes
## 12 3 4 0 gray83 0.5 12No
## 13 1 5 0 gray83 0.5 13No
## 14 2 5 0 gray83 0.5 14No
## 15 3 5 1 orange 1.0 5yes
## 16 1 6 0 gray83 0.5 16No
## 17 2 6 1 blue 1.0 6yes
## 18 3 6 0 gray83 0.5 18No
## 19 1 7 1 maroon 1.0 7yes
## 20 2 7 0 gray83 0.5 20No
## 21 3 7 0 gray83 0.5 21NoOk, let’s continue with the rest of upset().
## continuing with upset()
Set_sizes <- UpSetR:::FindSetFreqs(New_data, first.col, Num_of_set, Set_names, keep.order)
Bar_Q <- NULL
if(is.null(queries) == F){
Bar_Q <- UpSetR:::intersects(QuerieInterBar, Intersection, New_data, first.col, Num_of_set, All_Freqs, expression, Set_names, palette)
}
QInter_att_data <- NULL
QElem_att_data <- NULL
if((is.null(queries) == F) & (is.null(att.x) == F)){
QInter_att_data <- UpSetR:::intersects(QuerieInterAtt, Intersection, New_data, first.col, Num_of_set, att.x, att.y,
expression, Set_names, palette)
QElem_att_data <- UpSetR:::elements(QuerieElemAtt, Element, New_data, first.col, expression, Set_names, att.x, att.y,
palette)
}
AllQueryData <- UpSetR:::combineQueriesData(QInter_att_data, QElem_att_data, customAttDat, att.x, att.y)
ShadingData <- NULL
if(is.null(set.metadata) == F){
ShadingData <- get_shade_groups(set.metadata, Set_names, Matrix_layout, shade.alpha)
output <- Make_set_metadata_plot(set.metadata, Set_names)
set.metadata.plots <- output[[1]]
set.metadata <- output[[2]]
if(is.null(ShadingData) == FALSE){
shade.alpha <- unique(ShadingData$alpha)
}
} else {
set.metadata.plots <- NULL
}
if(is.null(ShadingData) == TRUE){
ShadingData <- UpSetR:::MakeShading(Matrix_layout, shade.color)
}
Main_bar <- suppressMessages(UpSetR:::Make_main_bar(All_Freqs, Bar_Q, show.numbers, mb.ratio, customQBar, number.angles, EBar_data, mainbar.y.label,
mainbar.y.max, scale.intersections, text.scale, attribute.plots))
Matrix <- UpSetR:::Make_matrix_plot(Matrix_layout, Set_sizes, All_Freqs, point.size, line.size,
text.scale, labels, ShadingData, shade.alpha)
Sizes <- UpSetR:::Make_size_plot(Set_sizes, sets.bar.color, mb.ratio, sets.x.label, scale.sets, text.scale, set_size.angles,set_size.show)
# Make_base_plot(Main_bar, Matrix, Sizes, labels, mb.ratio, att.x, att.y, New_data,
# expression, att.pos, first.col, att.color, AllQueryData, attribute.plots,
# legend, query.legend, BoxPlots, Set_names, set.metadata, set.metadata.plots)
structure(class = "upset",
.Data=list(
Main_bar = Main_bar,
Matrix = Matrix,
Sizes = Sizes,
labels = labels,
mb.ratio = mb.ratio,
att.x = att.x,
att.y = att.y,
New_data = New_data,
expression = expression,
att.pos = att.pos,
first.col = first.col,
att.color = att.color,
AllQueryData = AllQueryData,
attribute.plots = attribute.plots,
legend = legend,
query.legend = query.legend,
BoxPlots = BoxPlots,
Set_names = Set_names,
set.metadata = set.metadata,
set.metadata.plots = set.metadata.plots)
)
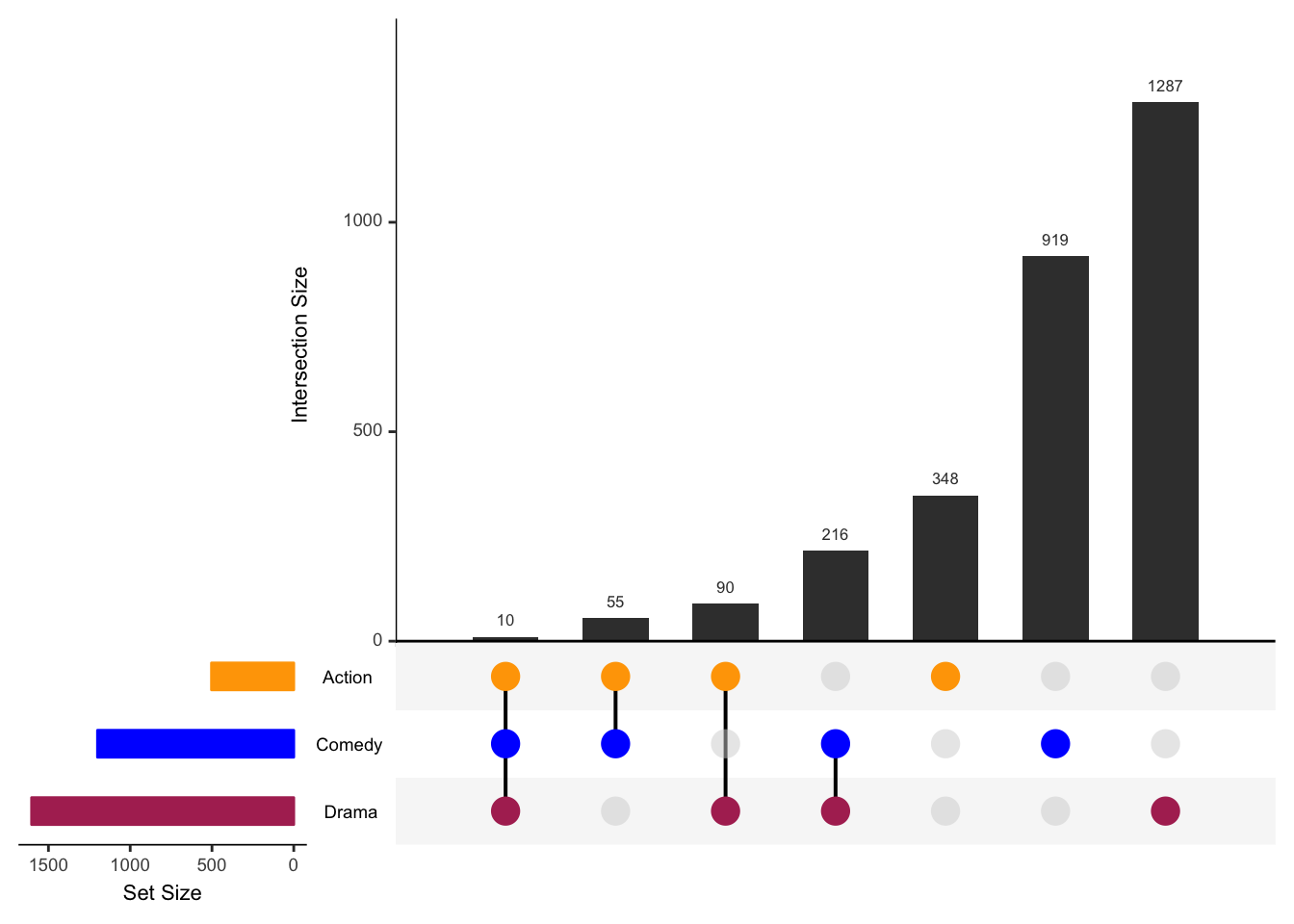
Line colors
Ok, that’s great but we have a problem with the lines. The color is no longer black, so we went deeper into the rabbit hole and found that the internal Make_matrix_plot() function is where the lines are made. We made some edits but got a plot where the lines were on top of the circles as shown in this screenshot.

Our club session was out of time, so we decided to continue our project another day and ask for help on twitter. And yay, we got help super fast!
Thank you and @thatdnaguy, that did it! pic.twitter.com/tzQvhKFXgR
— LIBD rstats club (@LIBDrstats) July 27, 2018
So here’s our modified version of Make_matrix_plot() that keeps the lines black.
Make_matrix_plot <- function(Mat_data,Set_size_data, Main_bar_data, point_size, line_size, text_scale, labels,
shading_data, shade_alpha){
if(length(text_scale) == 1){
name_size_scale <- text_scale
}
if(length(text_scale) > 1 && length(text_scale) <= 6){
name_size_scale <- text_scale[5]
}
Mat_data$line_col <- 'black'
Matrix_plot <- (ggplot()
+ theme(panel.background = element_rect(fill = "white"),
plot.margin=unit(c(-0.2,0.5,0.5,0.5), "lines"),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_text(colour = "gray0",
size = 7*name_size_scale, hjust = 0.4),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
+ xlab(NULL) + ylab(" ")
+ scale_y_continuous(breaks = c(1:nrow(Set_size_data)),
limits = c(0.5,(nrow(Set_size_data) +0.5)),
labels = labels, expand = c(0,0))
+ scale_x_continuous(limits = c(0,(nrow(Main_bar_data)+1 )), expand = c(0,0))
+ geom_rect(data = shading_data, aes_string(xmin = "min", xmax = "max",
ymin = "y_min", ymax = "y_max"),
fill = shading_data$shade_color, alpha = shade_alpha)
+ geom_line(data= Mat_data, aes_string(group = "Intersection", x="x", y="y",
colour = "line_col"), size = line_size)
+ geom_point(data= Mat_data, aes_string(x= "x", y= "y"), colour = Mat_data$color,
size= point_size, alpha = Mat_data$alpha, shape=16)
+ scale_color_identity())
Matrix_plot <- ggplot_gtable(ggplot_build(Matrix_plot))
return(Matrix_plot)
}Using that modified version we can then run the code again (note that we are not using UpSetR::: before Make_matrix_plot) and get the plot we wanted.
Matrix <- Make_matrix_plot(Matrix_layout, Set_sizes, All_Freqs, point.size, line.size,
text.scale, labels, ShadingData, shade.alpha)
Sizes <- UpSetR:::Make_size_plot(Set_sizes, sets.bar.color, mb.ratio, sets.x.label, scale.sets, text.scale, set_size.angles,set_size.show)
# Make_base_plot(Main_bar, Matrix, Sizes, labels, mb.ratio, att.x, att.y, New_data,
# expression, att.pos, first.col, att.color, AllQueryData, attribute.plots,
# legend, query.legend, BoxPlots, Set_names, set.metadata, set.metadata.plots)
structure(class = "upset",
.Data=list(
Main_bar = Main_bar,
Matrix = Matrix,
Sizes = Sizes,
labels = labels,
mb.ratio = mb.ratio,
att.x = att.x,
att.y = att.y,
New_data = New_data,
expression = expression,
att.pos = att.pos,
first.col = first.col,
att.color = att.color,
AllQueryData = AllQueryData,
attribute.plots = attribute.plots,
legend = legend,
query.legend = query.legend,
BoxPlots = BoxPlots,
Set_names = Set_names,
set.metadata = set.metadata,
set.metadata.plots = set.metadata.plots)
)
We have quite a bit more to do in order to complete our pull request. We are also curious if you would have used a different approach to hack your way through UpSetR (Gehlenborg, 2016). For example, maybe some functions from devtools (Wickham, Hester, and Chang, 2018) would have enabled to do this equally fast without having to introduce UpSetR::: calls.
Thanks for reading!
Acknowledgments
This blog post was made possible thanks to:
References
[1] C. Boettiger. knitcitations: Citations for ‘Knitr’ Markdown Files. R package version 1.0.8. 2017. URL: https://CRAN.R-project.org/package=knitcitations.
[2] N. Gehlenborg. UpSetR: A More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. R package version 1.4.0. 2016. URL: http://github.com/hms-dbmi/UpSetR.
[3] A. Oleś, M. Morgan and W. Huber. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.8.2. 2018. URL: https://github.com/Bioconductor/BiocStyle.
[4] H. Wickham, J. Hester and W. Chang. devtools: Tools to Make Developing R Packages Easier. R package version 1.13.6. 2018. URL: https://CRAN.R-project.org/package=devtools.
[5] Y. Xie, A. P. Hill and A. Thomas. blogdown: Creating Websites with R Markdown. ISBN 978-0815363729. Boca Raton, Florida: Chapman and Hall/CRC, 2017. URL: https://github.com/rstudio/blogdown.
Reproducibility
## Session info ----------------------------------------------------------------------------------------------------------## setting value
## version R version 3.5.1 (2018-07-02)
## system x86_64, darwin15.6.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz America/New_York
## date 2018-07-27## Packages --------------------------------------------------------------------------------------------------------------## package * version date source
## assertthat 0.2.0 2017-04-11 cran (@0.2.0)
## backports 1.1.2 2017-12-13 cran (@1.1.2)
## base * 3.5.1 2018-07-05 local
## bibtex 0.4.2 2017-06-30 CRAN (R 3.5.0)
## bindr 0.1.1 2018-03-13 cran (@0.1.1)
## bindrcpp 0.2.2 2018-03-29 cran (@0.2.2)
## BiocStyle * 2.8.2 2018-05-30 Bioconductor
## blogdown 0.8 2018-07-15 CRAN (R 3.5.0)
## bookdown 0.7 2018-02-18 CRAN (R 3.5.0)
## colorout * 1.2-0 2018-05-03 Github (jalvesaq/colorout@c42088d)
## colorspace 1.3-2 2016-12-14 cran (@1.3-2)
## compiler 3.5.1 2018-07-05 local
## crayon 1.3.4 2017-09-16 cran (@1.3.4)
## datasets * 3.5.1 2018-07-05 local
## devtools * 1.13.6 2018-06-27 cran (@1.13.6)
## digest 0.6.15 2018-01-28 CRAN (R 3.5.0)
## dplyr 0.7.6 2018-06-29 CRAN (R 3.5.1)
## evaluate 0.11 2018-07-17 CRAN (R 3.5.0)
## ggplot2 * 3.0.0 2018-07-03 CRAN (R 3.5.0)
## glue 1.3.0 2018-07-17 CRAN (R 3.5.0)
## graphics * 3.5.1 2018-07-05 local
## grDevices * 3.5.1 2018-07-05 local
## grid * 3.5.1 2018-07-05 local
## gridExtra * 2.3 2017-09-09 CRAN (R 3.5.0)
## gtable 0.2.0 2016-02-26 CRAN (R 3.5.0)
## htmltools 0.3.6 2017-04-28 cran (@0.3.6)
## httr 1.3.1 2017-08-20 CRAN (R 3.5.0)
## jsonlite 1.5 2017-06-01 CRAN (R 3.5.0)
## knitcitations * 1.0.8 2017-07-04 CRAN (R 3.5.0)
## knitr 1.20 2018-02-20 cran (@1.20)
## labeling 0.3 2014-08-23 cran (@0.3)
## lazyeval 0.2.1 2017-10-29 CRAN (R 3.5.0)
## lubridate 1.7.4 2018-04-11 CRAN (R 3.5.0)
## magrittr 1.5 2014-11-22 cran (@1.5)
## memoise 1.1.0 2017-04-21 CRAN (R 3.5.0)
## methods * 3.5.1 2018-07-05 local
## munsell 0.5.0 2018-06-12 CRAN (R 3.5.0)
## pillar 1.3.0 2018-07-14 CRAN (R 3.5.0)
## pkgconfig 2.0.1 2017-03-21 cran (@2.0.1)
## plyr * 1.8.4 2016-06-08 cran (@1.8.4)
## purrr 0.2.5 2018-05-29 cran (@0.2.5)
## R6 2.2.2 2017-06-17 CRAN (R 3.5.0)
## Rcpp 0.12.18 2018-07-23 CRAN (R 3.5.1)
## RefManageR 1.2.0 2018-04-25 CRAN (R 3.5.0)
## rlang 0.2.1 2018-05-30 cran (@0.2.1)
## rmarkdown 1.10 2018-06-11 CRAN (R 3.5.0)
## rprojroot 1.3-2 2018-01-03 cran (@1.3-2)
## scales 0.5.0 2017-08-24 cran (@0.5.0)
## stats * 3.5.1 2018-07-05 local
## stringi 1.2.4 2018-07-20 CRAN (R 3.5.0)
## stringr 1.3.1 2018-05-10 CRAN (R 3.5.0)
## tibble 1.4.2 2018-01-22 cran (@1.4.2)
## tidyselect 0.2.4 2018-02-26 cran (@0.2.4)
## tools 3.5.1 2018-07-05 local
## UpSetR * 1.4.0 2018-07-27 Github (hms-dbmi/UpSetR@fe2812c)
## utils * 3.5.1 2018-07-05 local
## withr 2.1.2 2018-03-15 CRAN (R 3.5.0)
## xfun 0.3 2018-07-06 CRAN (R 3.5.0)
## xml2 1.2.0 2018-01-24 CRAN (R 3.5.0)
## yaml 2.1.19 2018-05-01 CRAN (R 3.5.0)