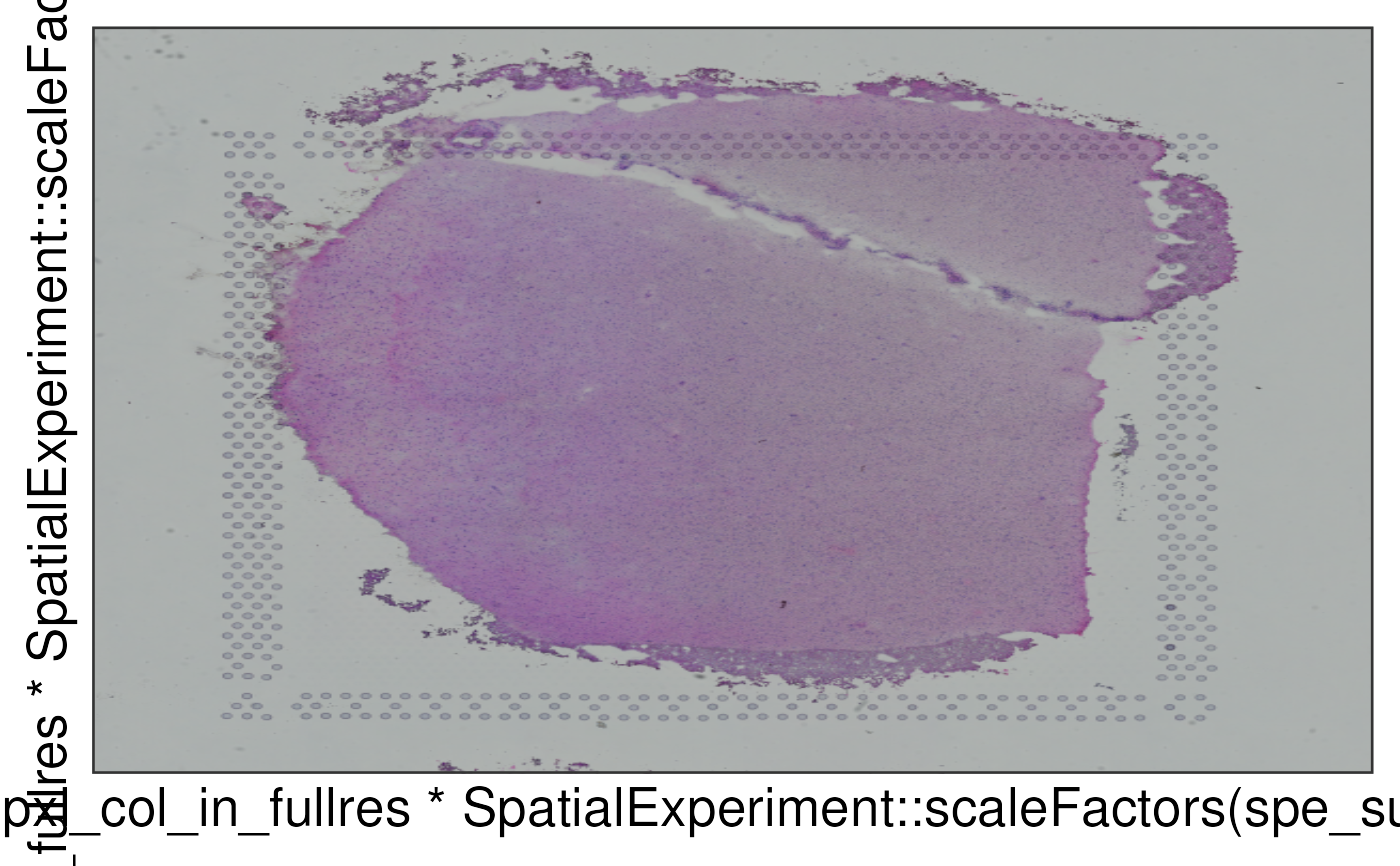
This function defines a ggplot2::layer() for visualizing the histology
image from Visium. It can be combined with other ggplot2 functions for
visualizing the clusters as in vis_clus_p() or gene-level information
as in vis_gene_p().
geom_spatial(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = FALSE,
...
)Arguments
- mapping
Passed to
ggplot2::layer(mapping)wheregrob,xandyare required.- data
Passed to
ggplot2::layer(data).- stat
Passed to
ggplot2::layer(stat).- position
Passed to
ggplot2::layer(position).- na.rm
Passed to
ggplot2::layer(params = list(na.rm)).- show.legend
Passed to
ggplot2::layer(show.legend).- inherit.aes
Passed to
ggplot2::layer(inherit.aes).- ...
Other arguments passed to
ggplot2::layer(params = list(...)).
Value
A ggplot2::layer() for the histology information.
Examples
if (enough_ram()) {
## Obtain the necessary data
if (!exists("spe")) spe <- fetch_data("spe")
## Select the first sample and extract the data
sample_id <- unique(spe$sample_id)[1]
spe_sub <- spe[, spe$sample_id == sample_id]
sample_df <- as.data.frame(colData(spe_sub), optional = TRUE)
## Obtain the histology image
img <- SpatialExperiment::imgRaster(spe_sub)
## Transform to a rasterGrob object
grob <- grid::rasterGrob(img, width = grid::unit(1, "npc"), height = grid::unit(1, "npc"))
## Make a plot using geom_spatial
p <- ggplot2::ggplot(
sample_df,
ggplot2::aes(
x = pxl_col_in_fullres * SpatialExperiment::scaleFactors(spe_sub),
y = pxl_row_in_fullres * SpatialExperiment::scaleFactors(spe_sub),
)
) +
geom_spatial(
data = tibble::tibble(grob = list(grob)),
ggplot2::aes(grob = grob),
x = 0.5,
y = 0.5
)
## Show the plot
print(p)
## Clean up
rm(spe_sub)
}
#> 2026-01-09 17:22:18.640718 loading file /github/home/.cache/R/BiocFileCache/1b5c76453a2f_Human_DLPFC_Visium_processedData_sce_scran_spatialLIBD.Rdata%3Fdl%3D1