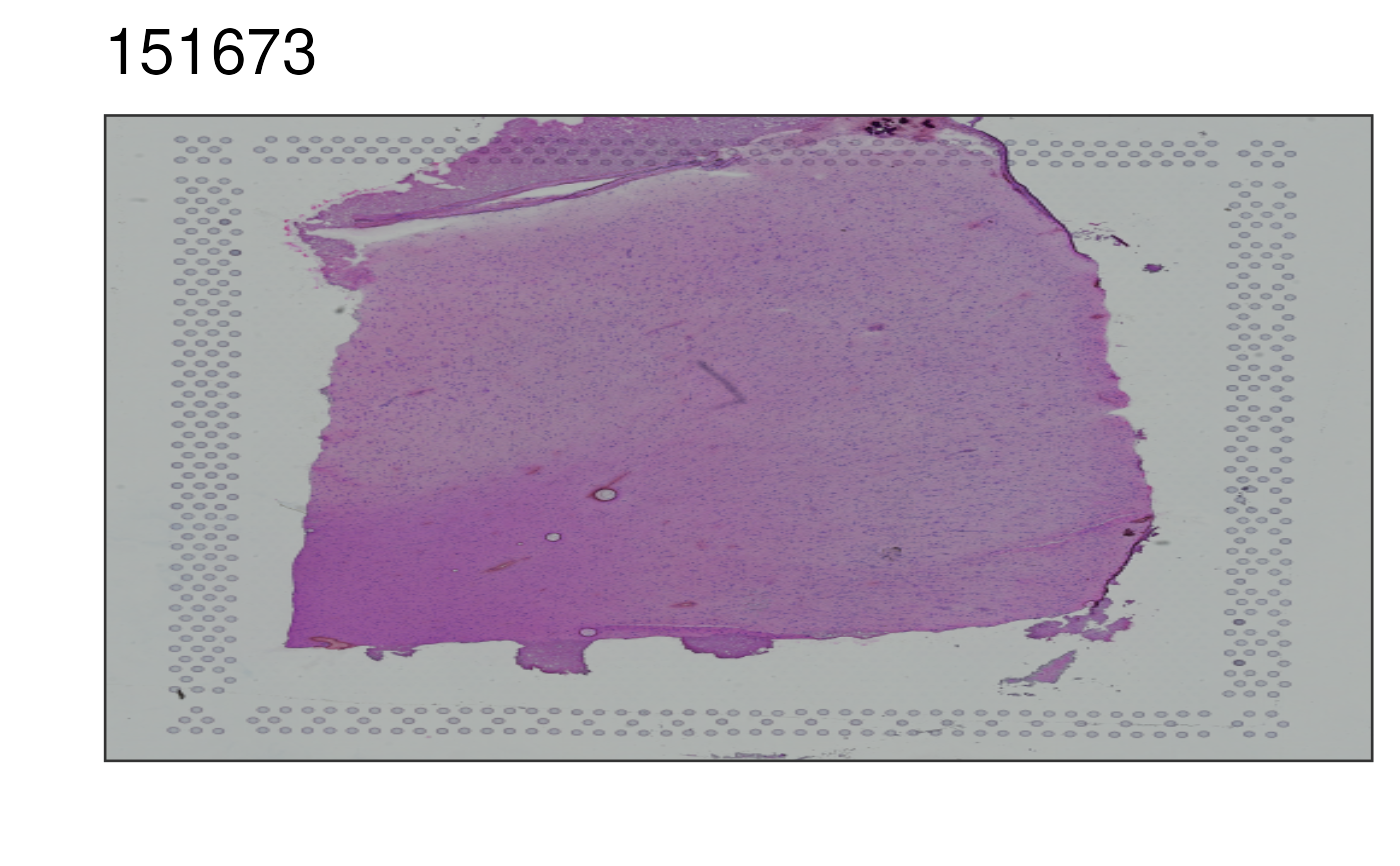
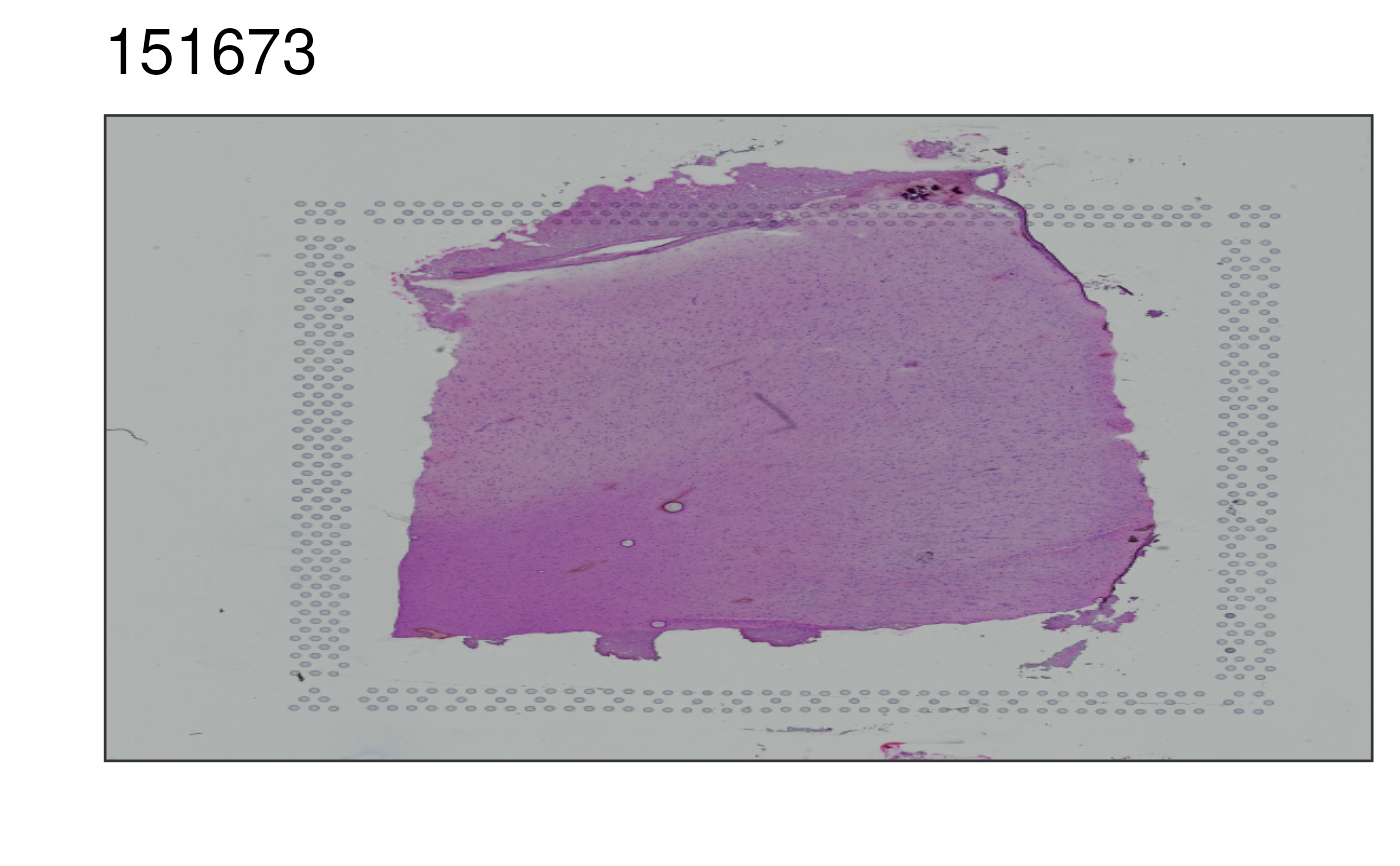
This function visualizes the histology image for selected sample. Matches
crop and settings of vis_clus() and vis_gene().
vis_image(
spe,
sampleid = unique(spe$sample_id)[1],
image_id = "lowres",
auto_crop = TRUE,
is_stitched = FALSE,
title_suffix = NULL
)Arguments
- spe
A SpatialExperiment-class object. See
fetch_data()for how to download some example objects orread10xVisiumWrapper()to read inspaceranger --countoutput files and build your ownspeobject.- sampleid
A
character(1)specifying which sample to plot fromcolData(spe)$sample_id(formerlycolData(spe)$sample_name).- image_id
A
character(1)with the name of the image ID you want to use in the background.- auto_crop
A
logical(1)indicating whether to automatically crop the image / plotting area, which is useful if the Visium capture area is not centered on the image and if the image is not a square.- is_stitched
A
logical(1)vector: IfTRUE, expects a SpatialExperiment-class built withvisiumStitched::build_spe(). http://research.libd.org/visiumStitched/reference/build_spe.html; in particular, expects a logical colData columnexclude_overlappingspecifying which spots to exclude from the plot. Setsauto_crop = FALSE.- title_suffix
A
character(1)passed to paste() to modify the title of the plot following thesampleid.
Value
A ggplot2 object.
Details
This function subsets spe to the given sample and prepares the
data and title for vis_clus_p().
See also
Other Spatial cluster visualization functions:
frame_limits(),
vis_clus(),
vis_clus_p(),
vis_grid_clus()
Examples
if (enough_ram()) {
## Obtain the necessary data
if (!exists("spe")) spe <- fetch_data("spe")
## Print the "lowres" image for sample 151673
p1 <- vis_image(
spe = spe,
sampleid = "151673"
)
print(p1)
## Without auto-cropping the image
p2 <- vis_image(
spe = spe,
sampleid = "151673",
auto_crop = FALSE
)
print(p2)
}
#> 2025-11-20 15:40:42.215805 loading file /github/home/.cache/R/BiocFileCache/184676c6874f_Human_DLPFC_Visium_processedData_sce_scran_spatialLIBD.Rdata%3Fdl%3D1