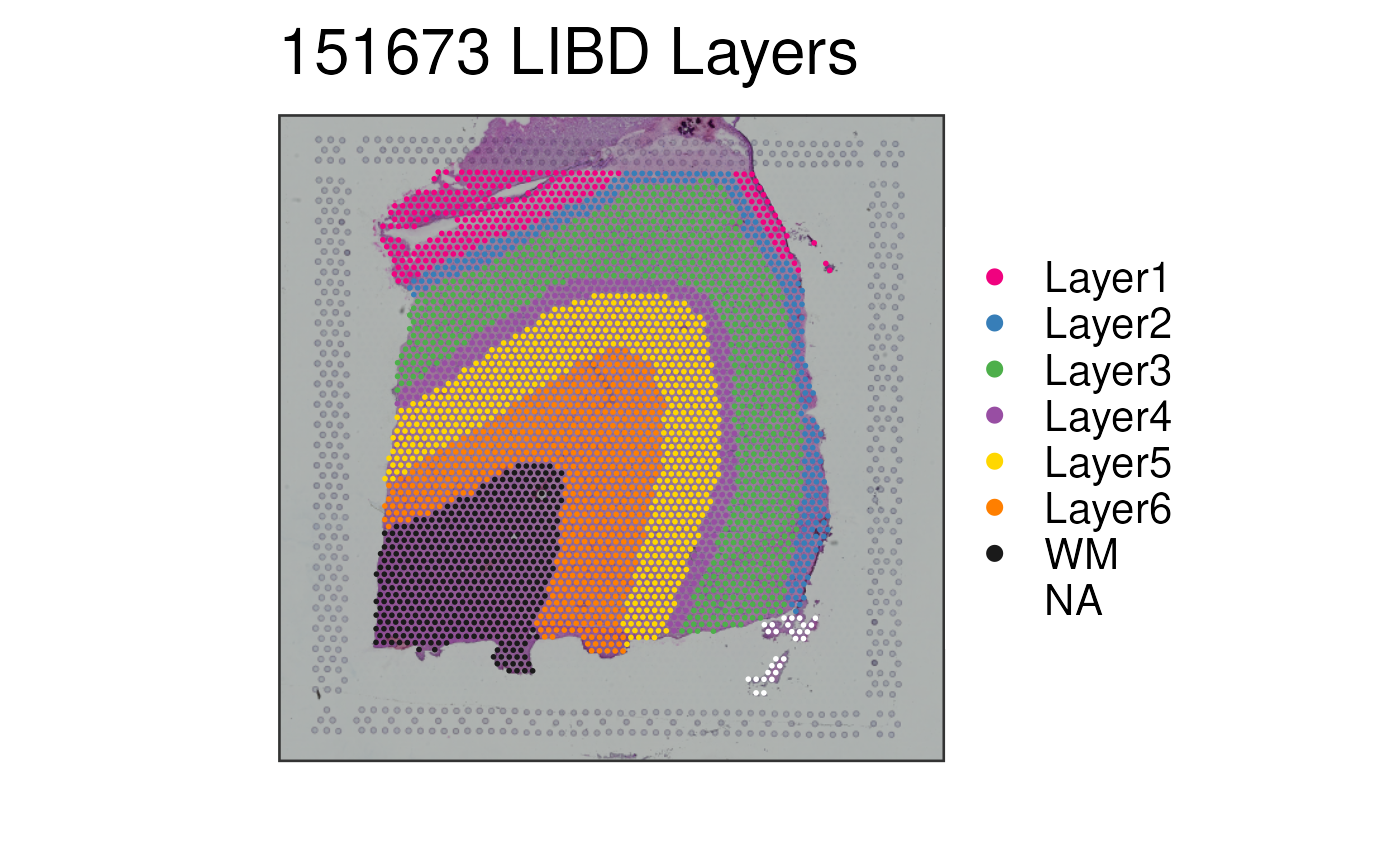
This function visualizes the clusters for one given sample at the spot-level
using (by default) the histology information on the background. To visualize
gene-level (or any continuous variable) use vis_gene().
vis_clus(
spe,
sampleid = unique(spe$sample_id)[1],
clustervar,
colors = c("#b2df8a", "#e41a1c", "#377eb8", "#4daf4a", "#ff7f00", "gold", "#a65628",
"#999999", "black", "grey", "white", "purple"),
spatial = TRUE,
image_id = "lowres",
alpha = NA,
point_size = 2,
auto_crop = TRUE,
na_color = "#CCCCCC40",
is_stitched = FALSE,
guide_point_size = point_size,
...
)Arguments
- spe
A SpatialExperiment-class object. See
fetch_data()for how to download some example objects orread10xVisiumWrapper()to read inspaceranger --countoutput files and build your ownspeobject.- sampleid
A
character(1)specifying which sample to plot fromcolData(spe)$sample_id(formerlycolData(spe)$sample_name).- clustervar
A
character(1)with the name of thecolData(spe)column that has the cluster values.- colors
A vector of colors to use for visualizing the clusters from
clustervar. If the vector has names, then those should match the values ofclustervar.- spatial
A
logical(1)indicating whether to include the histology layer fromgeom_spatial(). If you plan to use ggplotly() then it's best to set this toFALSE.- image_id
A
character(1)with the name of the image ID you want to use in the background.- alpha
A
numeric(1)in the[0, 1]range that specifies the transparency level of the data on the spots.- point_size
A
numeric(1)specifying the size of the points. Defaults to1.25. Some colors look better if you use2for instance.- auto_crop
A
logical(1)indicating whether to automatically crop the image / plotting area, which is useful if the Visium capture area is not centered on the image and if the image is not a square.- na_color
A
character(1)specifying a color for the NA values. If you setalpha = NAthen it's best to setna_colorto a color that has alpha blending already, which will make non-NA values pop up more and the NA values will show with a lighter color. This behavior is lost whenalphais set to a non-NAvalue.- is_stitched
A
logical(1)vector: IfTRUE, expects a SpatialExperiment-class built withvisiumStitched::build_spe(). http://research.libd.org/visiumStitched/reference/build_spe.html; in particular, expects a logical colData columnexclude_overlappingspecifying which spots to exclude from the plot. Setsauto_crop = FALSE.- guide_point_size
A
numeric(1)specifying the size of the points in guide. Defaults topoint_size. Increase to improve visability.- ...
Passed to paste0() for making the title of the plot following the
sampleid.
Value
A ggplot2 object.
Details
This function subsets spe to the given sample and prepares the
data and title for vis_clus_p().
See also
Other Spatial cluster visualization functions:
frame_limits(),
vis_clus_p(),
vis_grid_clus(),
vis_image()
Examples
if (enough_ram()) {
## Obtain the necessary data
if (!exists("spe")) spe <- fetch_data("spe")
## Check the colors defined by Lukas M Weber
libd_layer_colors
## Use the manual color palette by Lukas M Weber
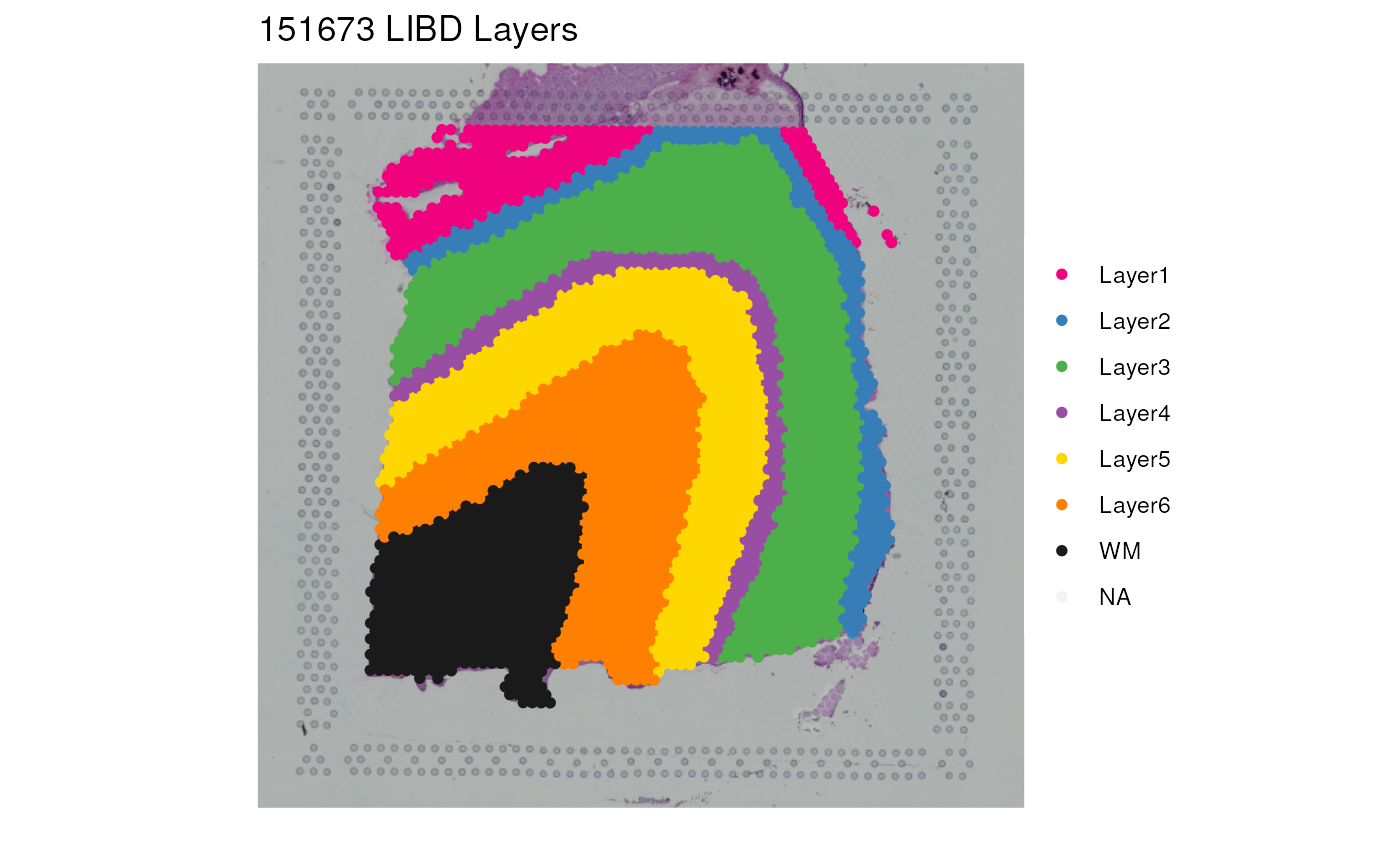
p1 <- vis_clus(
spe = spe,
clustervar = "layer_guess_reordered",
sampleid = "151673",
colors = libd_layer_colors,
... = " LIBD Layers"
)
print(p1)
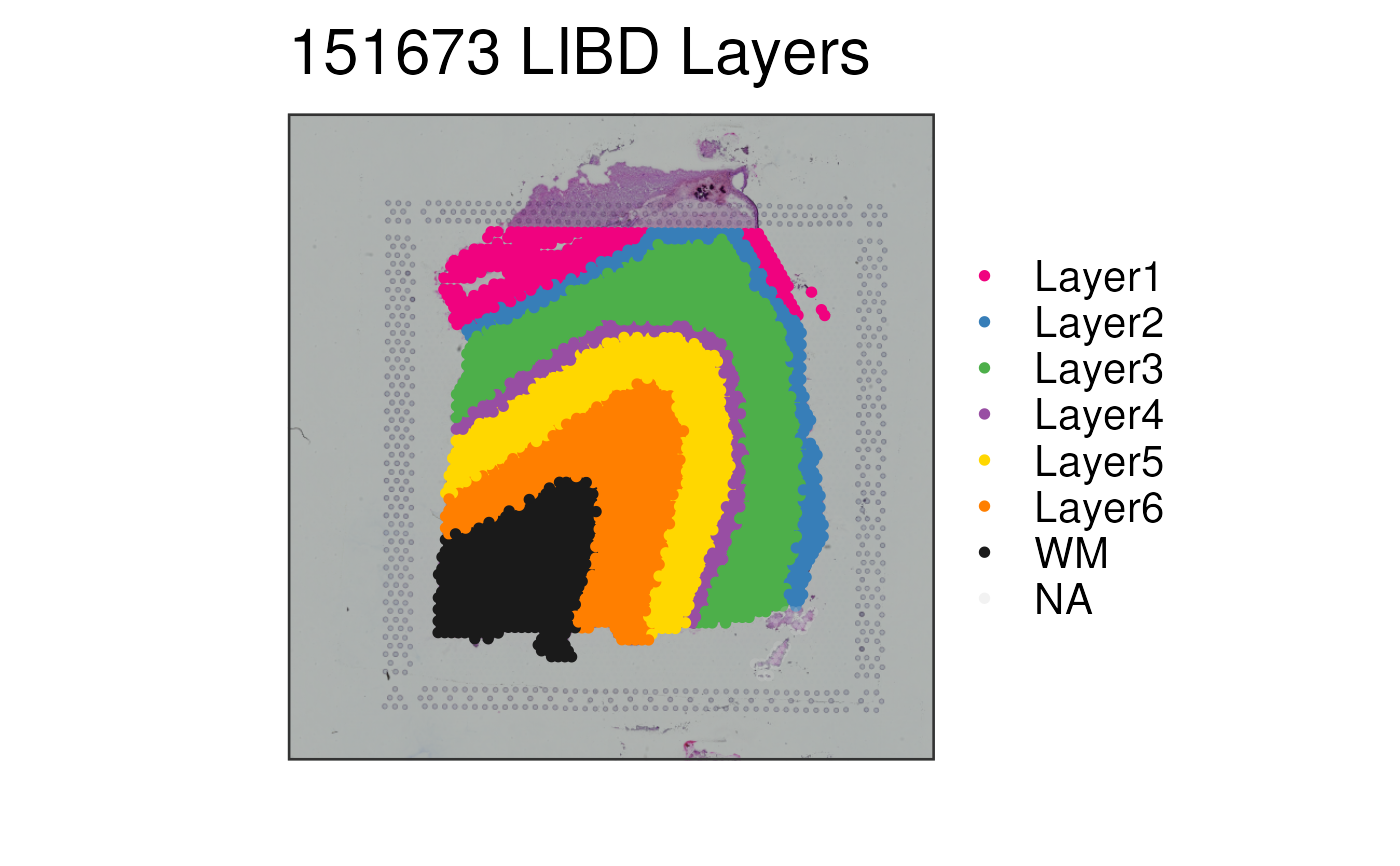
## Without auto-cropping the image
p2 <- vis_clus(
spe = spe,
clustervar = "layer_guess_reordered",
sampleid = "151673",
colors = libd_layer_colors,
auto_crop = FALSE,
... = " LIBD Layers"
)
print(p2)
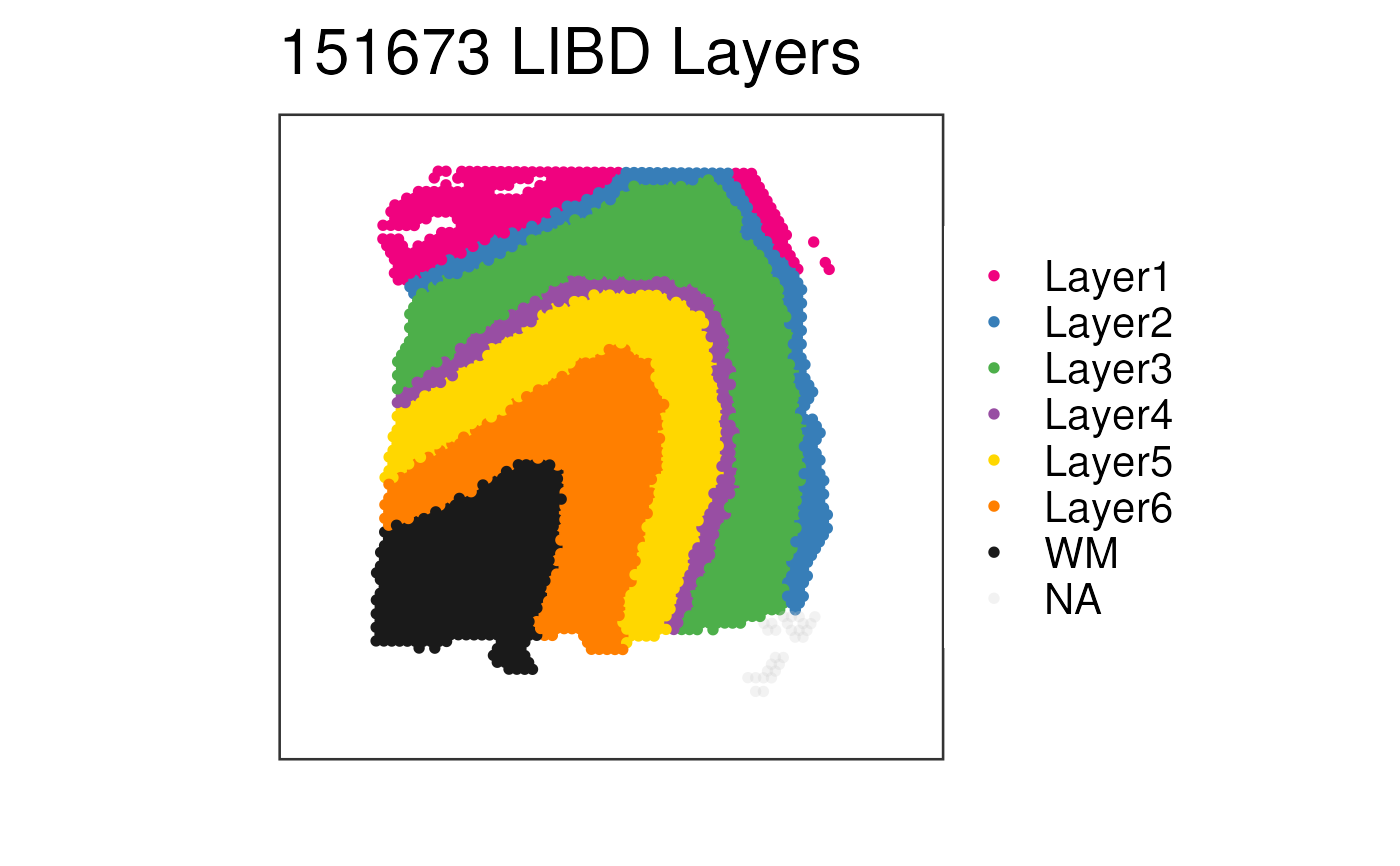
## Without histology
p3 <- vis_clus(
spe = spe,
clustervar = "layer_guess_reordered",
sampleid = "151673",
colors = libd_layer_colors,
... = " LIBD Layers",
spatial = FALSE
)
print(p3)
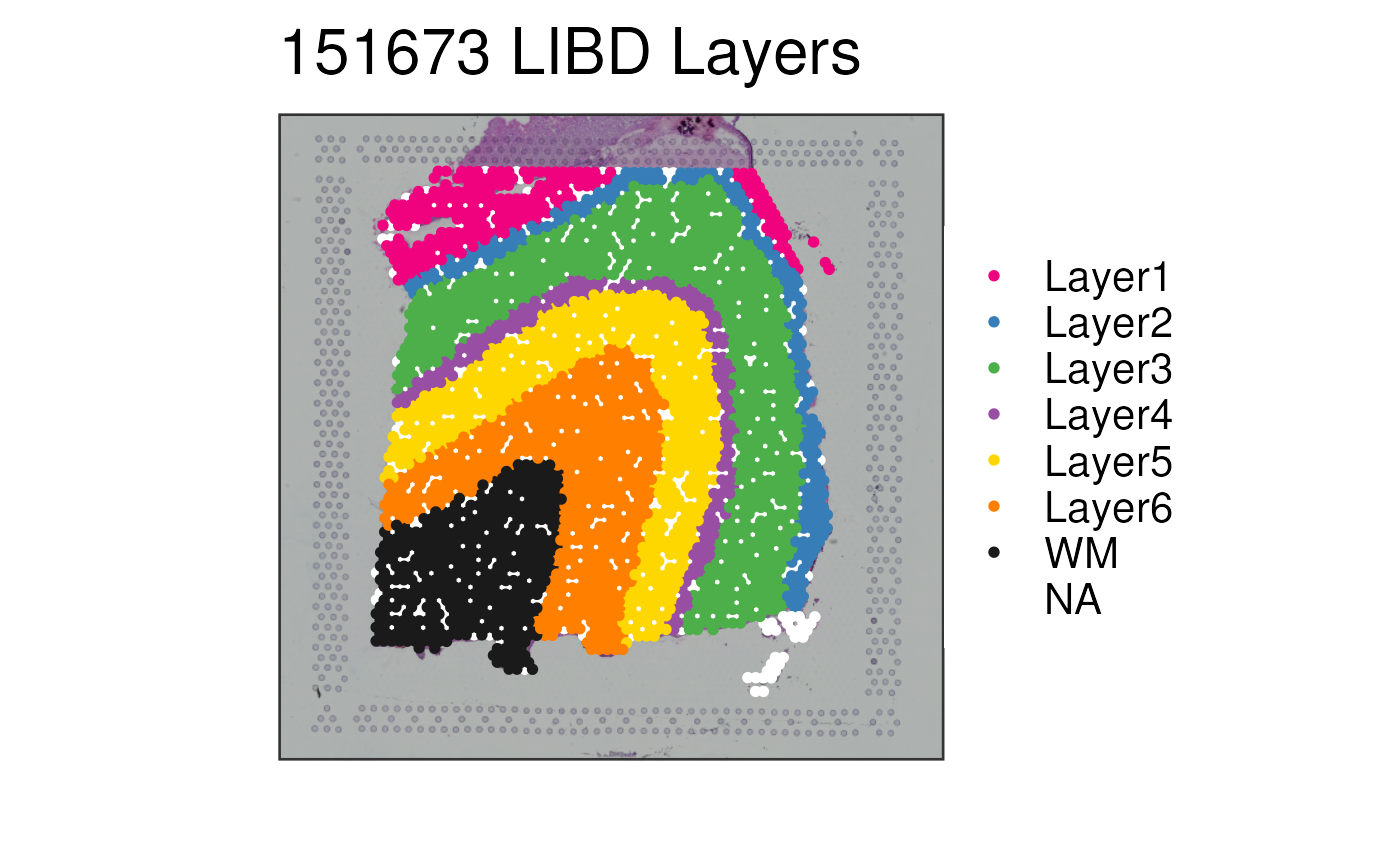
## With some NA values
spe$tmp <- spe$layer_guess_reordered
spe$tmp[spe$sample_id == "151673"][seq_len(500)] <- NA
p4 <- vis_clus(
spe = spe,
clustervar = "tmp",
sampleid = "151673",
colors = libd_layer_colors,
na_color = "white",
... = " LIBD Layers"
)
print(p4)
## edit plot point size but keep guide size larger
p5 <- vis_clus(
spe = spe,
clustervar = "layer_guess_reordered",
sampleid = "151673",
colors = libd_layer_colors,
na_color = "white",
point_size = 1,
guide_point_size = 3,
... = " LIBD Layers"
)
print(p5)
}
#> 2026-01-09 17:23:43.656438 loading file /github/home/.cache/R/BiocFileCache/1b5c76453a2f_Human_DLPFC_Visium_processedData_sce_scran_spatialLIBD.Rdata%3Fdl%3D1