Plot a nested list of genes as a multi-page pdf
Source:R/plot_marker_express_List.R
plot_marker_express_List.RdThis function plots a nested list of genes as a multi-page PDF, one for each sub list. A use case is plotting known marker genes for multiple cell types over cell type clusters with unknown identities.
plot_marker_express_List(
sce,
gene_list,
pdf_fn = NULL,
cellType_col = "cellType",
gene_name_col = "gene_name",
color_pal = NULL,
plot_points = FALSE
)Arguments
- sce
SummarizedExperiment-class object
- gene_list
A named
list()ofcharacter()vectors containing the names of genes to plot.- pdf_fn
A
character()of the pdf filename to plot to, ifNULLreturns all plots- cellType_col
The
character(1)name ofcolData()column containing cell type forscedata. It should matchcellType.targetinstats.- gene_name_col
The
character(1)name ofrowData()matching the gene name fromgene_list.- color_pal
A named
character(1)vector that contains a color palette matching thecell_typevalues.- plot_points
A
logical(1)indicating whether to plot points over the violin, defaults toFALSEas these often become over plotted and quite large (especially when saved as PDF).
Value
A PDF file with violin plots for the expression of top marker genes for all cell types.
See also
Other expression plotting functions:
plot_gene_express(),
plot_marker_express(),
plot_marker_express_ALL()
Examples
## Fetch sce example data
if (!exists("sce_DLPFC_example")) sce_DLPFC_example <- fetch_deconvo_data("sce_DLPFC_example")
#> 2026-03-03 16:13:59.274271 Access ExperimentHub EH9626
#> see ?DeconvoBuddies and browseVignettes('DeconvoBuddies') for documentation
#> loading from cache
## Create list-of-lists of genes to plot, names of sub-list become title of page
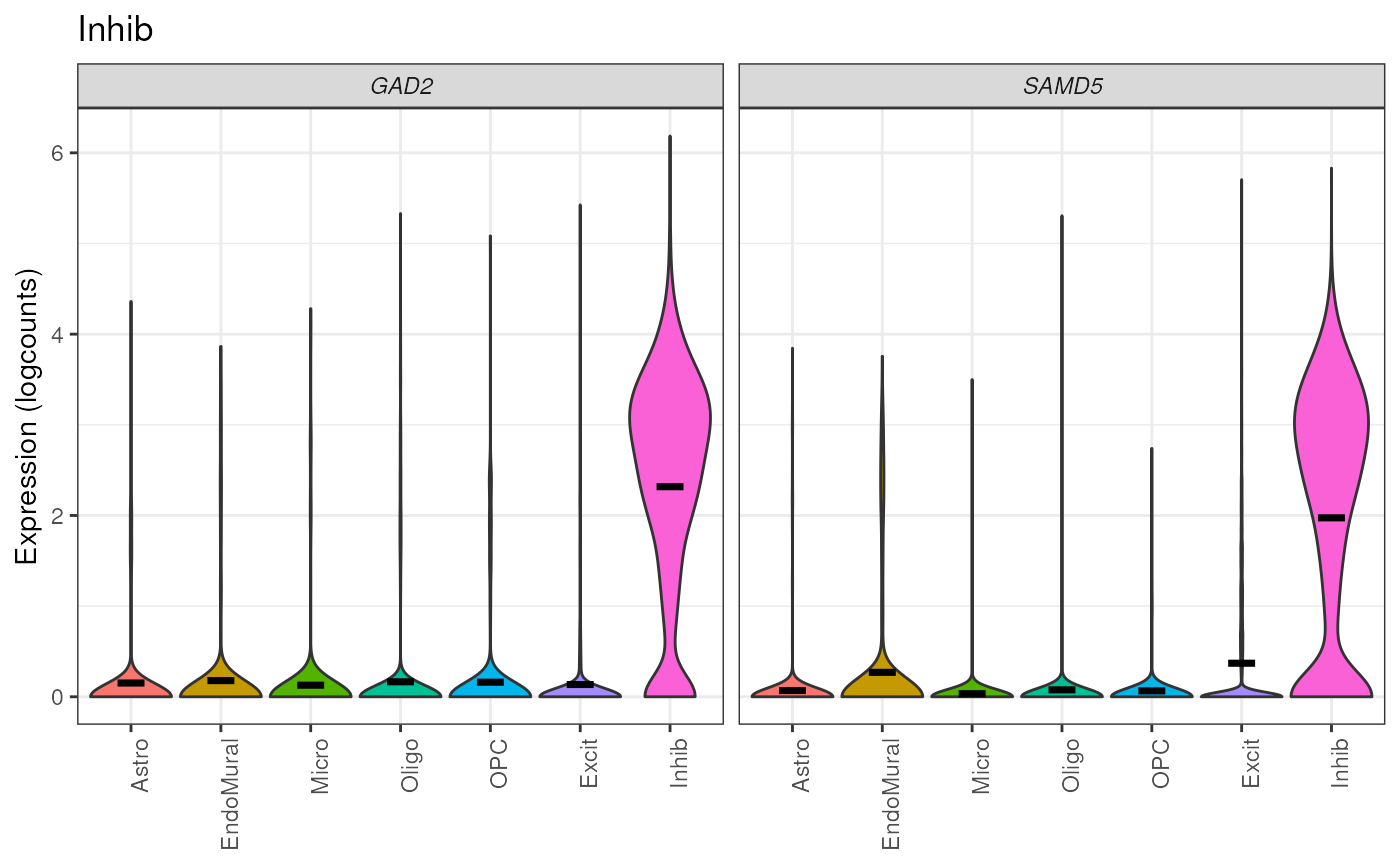
my_gene_list <- list(Inhib = c("GAD2", "SAMD5"), Astro = c("RGS20", "PRDM16"))
# Return a list of plots
plots <- plot_marker_express_List(
sce_DLPFC_example,
gene_list = my_gene_list,
cellType_col = "cellType_broad_hc"
)
print(plots[[1]])
 # Plot marker gene expression to PDF, one page per cell type in stats
pdf_file <- tempfile("test_marker_expression_List", fileext = ".pdf")
plot_marker_express_List(
sce_DLPFC_example,
gene_list = my_gene_list,
pdf_fn = pdf_file,
cellType_col = "cellType_broad_hc"
)
#> $Inhib
#>
#> $Astro
#>
#> agg_record_977209546
#> 2
if (interactive()) browseURL(pdf_file)
# Plot marker gene expression to PDF, one page per cell type in stats
pdf_file <- tempfile("test_marker_expression_List", fileext = ".pdf")
plot_marker_express_List(
sce_DLPFC_example,
gene_list = my_gene_list,
pdf_fn = pdf_file,
cellType_col = "cellType_broad_hc"
)
#> $Inhib
#>
#> $Astro
#>
#> agg_record_977209546
#> 2
if (interactive()) browseURL(pdf_file)